Protein Surface Interactions-Theoretical and Experimental Studies
- PMID: 34307462
- PMCID: PMC8298896
- DOI: 10.3389/fmolb.2021.706002
Protein Surface Interactions-Theoretical and Experimental Studies
Abstract
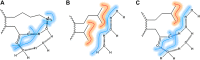
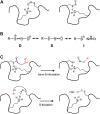
In this review, we briefly describe a theoretical discussion of protein folding, presenting the relative contribution of the hydrophobic effect versus the stabilization of proteins via direct surface forces that sometimes may be overlooked. We present NMR-based studies showing the stability of proteins lacking a hydrophobic core which in turn present hydrophobic surface clusters, such as plant defensins. Protein dynamics measurements by NMR are the key feature to understand these dynamic surface clusters. We contextualize the measurement of protein dynamics by nuclear relaxation and the information available at protein surfaces and water cavities. We also discuss the presence of hydrophobic surface clusters in multidomain proteins and their participation in transient interactions which may regulate the function of these proteins. In the end, we discuss how surface interaction regulates the reactivity of certain protein post-translational modifications, such as S-nitrosation.
Keywords: NMR; clusters; dynamics; hydrophobic surface clusters; interdomain; solvation; surface.
Copyright © 2021 Almeida, Sanches, Pinheiro-Aguiar, Almeida and Caruso.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Ben-Naim A. (2013). Water's Contribution in Providing strong Solvent-Induced Forces in Protein Folding. Eur. Phys. J. Spec. Top. 223, 927–946. 10.1140/epjst/e2013-01981-1 - DOI
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

