Development of a Rapid In Vitro Screening Assay Using Metabolic Inhibitors to Detect Highly Selective Anticancer Agents
- PMID: 34308064
- PMCID: PMC8296616
- DOI: 10.1021/acsomega.1c02203
Development of a Rapid In Vitro Screening Assay Using Metabolic Inhibitors to Detect Highly Selective Anticancer Agents
Abstract
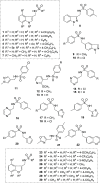
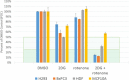
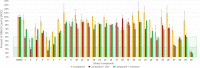
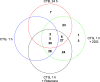
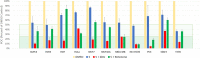
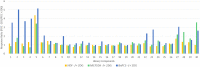
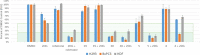
Traditional long exposure (24-72 h) cell viability assays for identification of potential drug compounds can fail to identify compounds that are: (a) biologically active but not toxic and (b) inactive without the addition of a synergistic additive. Herein, we report the development of a rapid (1-2 h) compound screening technique using a commercially available cell viability kit (CellTiter-Glo) that has led to the detection of compounds that were not identified as active agents using traditional cytotoxicity screening methods. These compounds, in combination with metabolic inhibitor 2-deoxyglucose, display selectivity toward a pancreatic cancer cell line. An evaluation of 11 mammalian cell lines against 30 novel compounds and two metabolic inhibitors is reported. The inclusion of metabolic inhibitors during an initial screening process, and not simply during mechanistic investigations of a previously identified hit compound, provides a rapid and sensitive tool for identifying drug candidates potentially overlooked by other methods.
© 2021 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



References
-
- American Cancer Society. Cancer Facts and Figures 2021. https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-... (accessed March 29, 2021).
-
- Hsu P. P.; Sabatini D. M. Cancer cell metabolism: Warburg and beyond. Cell 2008, 134, 703–707. 10.1016/j.cell.2008.08.021. - DOI - PubMed
- Resendis-Antonio O.; Checa A.; Encarnacion S. Modeling core metabolism in cancer cells: surveying the topology underlying the Warburg effect. PLoS One 2010, 5, e1238310.1371/journal.pone.0012383. - DOI - PMC - PubMed
- Flaveny C. A.; Griffett K.; El-Gendy Bel D.; Kazantzis M.; Sengupta M.; Amelio A. L.; Chatterjee A.; Walker J.; Solt L. A.; Kamenecka T. M.; Burris T. P. Broad Anti-tumor Activity of a Small Molecule that Selectively Targets the Warburg Effect and Lipogenesis. Cancer Cell 2015, 28, 42–56. 10.1016/j.ccell.2015.05.007. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

