In Situ Intraepithelial Localizations of Opportunistic Pathogens, Porphyromonas gingivalis and Filifactor alocis, in Human Gingiva
- PMID: 34308393
- PMCID: PMC8294339
- DOI: 10.1016/j.crmicr.2020.05.001
In Situ Intraepithelial Localizations of Opportunistic Pathogens, Porphyromonas gingivalis and Filifactor alocis, in Human Gingiva
Abstract
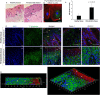
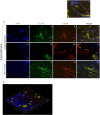
Porphyromonas gingivalis and Filifactor alocis are fastidious oral pathogens and etiological agents associated with chronic periodontitis. Although previous studies showed increased levels of the two obligate anaerobic species in periodontitis patients, methodologies for this knowledge were primarily limited to sampling subgingival plaque, saliva, or gingival crevicular fluid. To evaluate the extent to which P. gingivalis and F. alocis may invade the periodontal tissues, an in situ cross-sectional study was comparatively conducted on the gingival biopsy specimens of patients diagnosed with periodontal health or chronic periodontitis. Immunostained tissue sections for each organism were imaged by Super-Resolution Confocal Scanning Microscopy to determine the relative presence and localization of target bacterial species. Fluorescence-in-situ-hybridization (FISH) coupled with species specific 16S rRNA method was utilized to confirm whether detected bacteria were live within the tissue. In periodontitis, P. gingivalis and F. alocis revealed similarly concentrated localization near the basement membrane or external basal lamina of the gingival epithelium. The presence of both bacteria was significantly increased in periodontitis vs. healthy tissue. However, P. gingivalis was still detected to an extent in health tissue, while only minimal levels of F. alocis were spotted in health. Additionally, the micrographic analyses displayed heightened formation of epithelial microvasculature containing significantly co-localized and metabolically active dual species within periodontitis tissue. Thus, this study demonstrates, for the first-time, spatial arrangements of P. gingivalis and F. alocis in both single and co-localized forms within the complex fabric of human gingiva during health and disease. It also exhibits critical visualizations of co-invaded microvascularized epithelial layer of the tissue by metabolically active P. gingivalis and F. alocis from patients with severe periodontitis. These findings collectively uncover novel visual evidence of a potential starting point for systemic spread of opportunistic bacteria during their chronic colonization in gingival epithelium.
Keywords: Dual species colonization in gingiva; Filifactor alocis; Metabolically active bacteria in human tissue; Periodontal epithelium; Porphyromonas gingivalis.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




References
-
- American Academy of Periodontology Task Force Report on the Update to the 1999 Classification of Periodontal Diseases and Conditions. J Periodontol. 2015;86:835–838. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

