High-risk clones of extended-spectrum β-lactamase-producing Klebsiella pneumoniae isolated from the University Hospital Establishment of Oran, Algeria (2011-2012)
- PMID: 34310625
- PMCID: PMC8312963
- DOI: 10.1371/journal.pone.0254805
High-risk clones of extended-spectrum β-lactamase-producing Klebsiella pneumoniae isolated from the University Hospital Establishment of Oran, Algeria (2011-2012)
Abstract
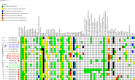
The purpose of the study was to characterize the resistome, virulome, mobilome and Clustered Regularly Interspaced Short Palindromic Repeats-associated (CRISPR-Cas) system of extended-spectrum β-lactamase producing Klebsiella pneumoniae (ESBL-KP) clinical isolates and to determine their phylogenetic relatedness. The isolates were from Algeria, isolated at the University Hospital Establishment of Oran, between 2011 and 2012. ESBL-KP isolates (n = 193) were screened for several antibiotic resistance genes (ARGs) using qPCR followed by Pulsed-Field Gel Electrophoresis (PFGE). Representative isolates were selected from PFGE clusters and subjected to whole-genome sequencing (WGS). Genomic characterization of the WGS data by studying prophages, CRISPR-Cas systems, Multi-Locus Sequence Typing (MLST), serotype, ARGs, virulence genes, plasmid replicons, and their pMLST. Phylogenetic and comparative genomic were done using core genome MLST and SNP-Based analysis. Generally, the ESBL-KP isolates were polyclonal. The whole genome sequences of nineteen isolates were taken of main PFGE clusters. Sixteen sequence types (ST) were found including high-risk clones ST14, ST23, ST37, and ST147. Serotypes K1 (n = 1), K2 (n = 2), K3 (n = 1), K31 (n = 1), K62 (n = 1), and K151 (n = 1) are associated with hyper-virulence. CRISPR-Cas system was found in 47.4%, typed I-E and I-E*. About ARGs, from 193 ESBL-KP, the majority of strains were multidrug-resistant, the CTX-M-1 enzyme was predominant (99%) and the prevalence of plasmid-mediated quinolone resistance (PMQR) genes was high with aac(6')-lb-cr (72.5%) and qnr's (65.8%). From 19 sequenced isolates we identified ESBL, AmpC, and carbapenemase genes: blaCTX-M-15 (n = 19), blaOXA-48 (n = 1), blaCMY-2 (n = 2), and blaCMY-16 (n = 2), as well as non-ESBL genes: qnrB1 (n = 12), qnrS1 (n = 1) and armA (n = 2). We found IncF, IncN, IncL/M, IncA/C2, and Col replicon types, at least once per isolate. This study is the first to report qnrS in ESBL-KP in Algeria. Our analysis shows the concerning co-existence of virulence and resistance genes and would support that genomic surveillance should be a high priority in the hospital environment.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- World Health Organization. Antimicrobial resistance: global report on surveillance. 2014. Available from: https://apps.who.int/iris/handle/10665/112642.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

