DNA methylome in visceral adipose tissue can discriminate patients with and without colorectal cancer
- PMID: 34311674
- PMCID: PMC9235880
- DOI: 10.1080/15592294.2021.1950991
DNA methylome in visceral adipose tissue can discriminate patients with and without colorectal cancer
Abstract
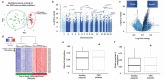
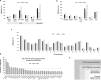
Adipose tissue dysfunction, particularly the visceral (VAT) compartment, has been proposed to play a relevant role in colorectal cancer (CRC) development and progression. Epigenetic mechanisms could be involved in this association. The current study aimed to evaluate if specific epigenetic marks in VAT are associated with colorectal cancer (CRC) to identify epigenetic hallmarks of adipose tissue-related CRC. Epigenome-wide DNA methylation was evaluated in VAT from 25 healthy participants and 29 CRC patients, using the Infinium HumanMethylation450K BeadChip. The epigenome-wide methylation analysis identified 170,184 sites able to perfectly separate the CRC and healthy samples. The differentially methylated CpG sites (DMCpGs) showed a global trend for increased methylated levels in CRC with respect to healthy group. Most of the genes encoded by the DMCpGs belonged to metabolic pathways and cell cycle, insulin resistance, and adipocytokine signalling, as well as tumoural transformation processes. In gene-specific analyses, involved genes biologically relevant for the development of CRC include PTPRN2, MAD1L1, TNXB, DIP2C, INPP5A, HDCA4, PRDM16, RPTOR, ATP11A, TBCD, PABPC3, and IER2. The methylation level of some of them showed a discriminatory capacity for detecting CRC higher than 90%, showing IER2 to have the highest capacity. This study reveals that a specific methylation pattern of VAT is associated with CRC. Some of the epigenetic marks identified could provide useful tools for the prediction and personalized treatment of CRC connected to excess adiposity.
Keywords: DNA methylation; adipose tissue; cancer; microarray; obesity.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures


References
-
- Wang YX, Zhu N, Zhang CJ, et al. Friend or foe: multiple roles of adipose tissue in cancer formation and progression. J Cell Physiol. 2019;234:21436–21449. - PubMed
-
- Crujeiras AB, Cabia B, Carreira MC, et al. Secreted factors derived from obese visceral adipose tissue regulate the expression of breast malignant transformation genes. Int J Obes. 2016;40(3):514–523. . - PubMed
-
- Spyrou N, Avgerinos KI, Mantzoros CS, et al. Classic and Novel Adipocytokines at the Intersection of Obesity and Cancer: diagnostic and Therapeutic Strategies. Curr Obes Rep. 2018;7:260–275. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
