Heterogeneity and Cancer-Related Features in Lymphangioleiomyomatosis Cells and Tissue
- PMID: 34312290
- PMCID: PMC8568632
- DOI: 10.1158/1541-7786.MCR-21-0220
Heterogeneity and Cancer-Related Features in Lymphangioleiomyomatosis Cells and Tissue
Abstract
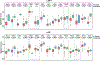
Lymphangioleiomyomatosis (LAM) is a rare, low-grade metastasizing disease characterized by cystic lung destruction. LAM can exhibit extensive heterogeneity at the molecular, cellular, and tissue levels. However, the molecular similarities and differences among LAM cells and tissue, and their connection to cancer features are not fully understood. By integrating complementary gene and protein LAM signatures, and single-cell and bulk tissue transcriptome profiles, we show sources of disease heterogeneity, and how they correspond to cancer molecular portraits. Subsets of LAM diseased cells differ with respect to gene expression profiles related to hormones, metabolism, proliferation, and stemness. Phenotypic diseased cell differences are identified by evaluating lumican (LUM) proteoglycan and YB1 transcription factor expression in LAM lung lesions. The RUNX1 and IRF1 transcription factors are predicted to regulate LAM cell signatures, and both regulators are expressed in LAM lung lesions, with differences between spindle-like and epithelioid LAM cells. The cancer single-cell transcriptome profiles most similar to those of LAM cells include a breast cancer mesenchymal cell model and lines derived from pleural mesotheliomas. Heterogeneity is also found in LAM lung tissue, where it is mainly determined by immune system factors. Variable expression of the multifunctional innate immunity protein LCN2 is linked to disease heterogeneity. This protein is found to be more abundant in blood plasma from LAM patients than from healthy women. IMPLICATIONS: This study identifies LAM molecular and cellular features, master regulators, cancer similarities, and potential causes of disease heterogeneity.
©2021 American Association for Cancer Research.
Conflict of interest statement
Figures

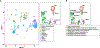

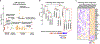

References
-
- Johnson SR, Cordier JF, Lazor R, Cottin V, Costabel U, Harari S, et al. European Respiratory Society guidelines for the diagnosis and management of lymphangioleiomyomatosis. Eur Respir J 2010;35:14–26. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

