Immunogenicity and reactogenicity of heterologous ChAdOx1 nCoV-19/mRNA vaccination
- PMID: 34312554
- PMCID: PMC8440177
- DOI: 10.1038/s41591-021-01464-w
Immunogenicity and reactogenicity of heterologous ChAdOx1 nCoV-19/mRNA vaccination
Abstract
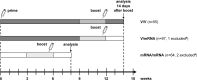
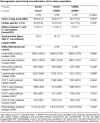
Heterologous priming with the ChAdOx1 nCoV-19 vector vaccine followed by boosting with a messenger RNA vaccine (BNT162b2 or mRNA-1273) is currently recommended in Germany, although data on immunogenicity and reactogenicity are not available. In this observational study we show that, in healthy adult individuals (n = 96), the heterologous vaccine regimen induced spike-specific IgG, neutralizing antibodies and spike-specific CD4 T cells, the levels of which which were significantly higher than after homologous vector vaccine boost (n = 55) and higher or comparable in magnitude to homologous mRNA vaccine regimens (n = 62). Moreover, spike-specific CD8 T cell levels after heterologous vaccination were significantly higher than after both homologous regimens. Spike-specific T cells were predominantly polyfunctional with largely overlapping cytokine-producing phenotypes in all three regimens. Recipients of both the homologous vector regimen and the heterologous vector/mRNA combination reported greater reactogenicity following the priming vector vaccination, whereas heterologous boosting was well tolerated and comparable to homologous mRNA boosting. Taken together, heterologous vector/mRNA boosting induces strong humoral and cellular immune responses with acceptable reactogenicity profiles.
© 2021. The Author(s).
Conflict of interest statement
M.S. has received grant support from Astellas and Biotest to the organization Saarland University outside the submitted work, and honoraria for lectures from Biotest and Novartis. All other authors of this paper have no conflicts of interest.
Figures




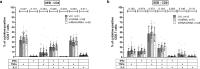

Comment in
-
A 'mix and match' approach to SARS-CoV-2 vaccination.Nat Med. 2021 Sep;27(9):1510-1511. doi: 10.1038/s41591-021-01463-x. Nat Med. 2021. PMID: 34312555 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

