Genetic Exchange of Lung-Derived Exosome to Brain Causing Neuronal Changes on COVID-19 Infection
- PMID: 34312772
- PMCID: PMC8313419
- DOI: 10.1007/s12035-021-02485-9
Genetic Exchange of Lung-Derived Exosome to Brain Causing Neuronal Changes on COVID-19 Infection
Abstract
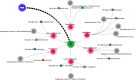
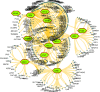
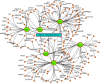
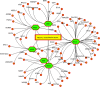
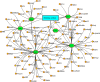
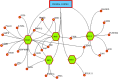
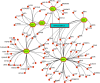
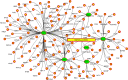
The pandemic of novel coronavirus 2 (SARS-CoV-2) has made global chaos for normal human living. Despite common COVID-19 symptoms, variability in clinical phenotypes was reported worldwide. Reports on SARS-CoV-2 suggest causing neurological manifestation. In addition, the susceptibility of SARS-CoV-2 in patients with neurodegenerative diseases and its complexity are largely unclear. Here, we aimed to demonstrate the possible transport of exosome from SARS-CoV-2-infected lungs to the brain regions associated with neurodegenerative diseases using multiple transcriptome datasets of SARS-CoV-2-infected lungs, RNA profiles from lung exosome, and gene expression profiles of the human brain. Upon transport, the transcription factors localized in the exosome regulate genes at lateral substantia nigra, medial substantia nigra, and superior frontal gyrus regions of Parkinson's disease (PD) and frontal cortex, hippocampus, and temporal cortex of Alzheimer's disease (AD). On SARS-CoV-2 infection, BCL3, JUND, MXD1, IRF2, IRF9, and STAT1 transcription factors in the exosomes influence the neuronal gene regulatory network and accelerate neurodegeneration. STAT1 transcription factor regulates 64 PD genes at lateral substantia nigra, 65 at superior frontal gyrus, and 19 at medial substantia nigra. Similarly, in AD, STAT1 regulates 74 AD genes at the temporal cortex, 40 genes at the hippocampus, and 16 genes at the frontal cortex. We further demonstrate that dysregulated neuronal genes showed involvement in immune response, signal transduction, apoptosis, and stress response process. In conclusion, SARS-CoV-2 may dysregulate neuronal gene regulatory network through exosomes that attenuate disease severity of neurodegeneration.
Keywords: Alzheimer’s disease; Covid-19; Exosome; Neurodegeneration; Parkinson’s disease; SARS‐CoV‐2.
© 2021. The Author(s), under exclusive licence to Springer Science+Business Media, LLC, part of Springer Nature.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Statello L, Maugeri M, Garre E, Nawaz M, Wahlgren J, Papadimitriou A, Lundqvist C, Lindfors L, et al. Identification of RNA-binding proteins in exosomes capable of interacting with different types of RNA: RBP-facilitated transport of RNAs into exosomes. PLoS One. 2018;13(4):e0195969. doi: 10.1371/journal.pone.0195969. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

