Marginating transitional B cells modulate neutrophils in the lung during inflammation and pneumonia
- PMID: 34313733
- PMCID: PMC8318832
- DOI: 10.1084/jem.20210409
Marginating transitional B cells modulate neutrophils in the lung during inflammation and pneumonia
Abstract
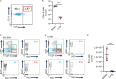
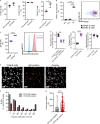
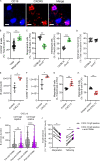
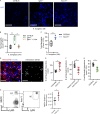
Pulmonary innate immunity is required for host defense; however, excessive neutrophil inflammation can cause life-threatening acute lung injury. B lymphocytes can be regulatory, yet little is known about peripheral transitional IgM+ B cells in terms of regulatory properties. Using single-cell RNA sequencing, we discovered eight IgM+ B cell subsets with unique gene regulatory networks in the lung circulation dominated by transitional type 1 B and type 2 B (T2B) cells. Lung intravital confocal microscopy revealed that T2B cells marginate in the pulmonary capillaries via CD49e and require CXCL13 and CXCR5. During lung inflammation, marginated T2B cells dampened excessive neutrophil vascular inflammation via the specialized proresolving molecule lipoxin A4 (LXA4). Exogenous CXCL13 dampened excessive neutrophilic inflammation by increasing marginated B cells, and LXA4 recapitulated neutrophil regulation in B cell-deficient mice during inflammation and fungal pneumonia. Thus, the lung microvasculature is enriched in multiple IgM+ B cell subsets with marginating capillary T2B cells that dampen neutrophil responses.
© 2021 Podstawka et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures






Comment in
-
T2B or not to B: Calming neutrophils offshore.J Exp Med. 2021 Sep 6;218(9):e20211407. doi: 10.1084/jem.20211407. Epub 2021 Aug 10. J Exp Med. 2021. PMID: 34374712 Free PMC article.
References
-
- Barletta, K.E., Cagnina R.E., Wallace K.L., Ramos S.I., Mehrad B., and Linden J.. 2012. Leukocyte compartments in the mouse lung: distinguishing between marginated, interstitial, and alveolar cells in response to injury. J. Immunol. Methods. 375:100–110. 10.1016/j.jim.2011.09.013 - DOI - PMC - PubMed
-
- Barnes, B.J., Adrover J.M., Baxter-Stoltzfus A., Borczuk A., Cools-Lartigue J., Crawford J.M., Daßler-Plenker J., Guerci P., Huynh C., Knight J.S., et al. . 2020. Targeting potential drivers of COVID-19: Neutrophil extracellular traps. J. Exp. Med. 217:e20200652. 10.1084/jem.20200652 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

