Adverse COVID-19 outcomes in immune deficiencies: Inequality exists between subclasses
- PMID: 34314546
- PMCID: PMC8441734
- DOI: 10.1111/all.15025
Adverse COVID-19 outcomes in immune deficiencies: Inequality exists between subclasses
Abstract
Background: Genetic deficiencies of immune system, referred to as inborn errors of immunity (IEI), serve as a valuable model to study human immune responses. In a multicenter prospective cohort, we evaluated the outcome of SARS-CoV-2 infection among IEI subjects and analyzed genetic and immune characteristics that determine adverse COVID-19 outcomes.
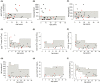
Methods: We studied 34 IEI patients (19M/15F, 12 [min: 0.6-max: 43] years) from six centers. We diagnosed COVID-19 infection by finding a positive SARS-CoV-2 PCR test (n = 25) and/or a lung tomography scoring (CORADS) ≥4 (n = 9). We recorded clinical and laboratory findings prospectively, fitted survival curves, and calculated fatality rates for the entire group and each IEI subclass.
Results: Nineteen patients had combined immune deficiency (CID), six with predominantly antibody deficiency (PAD), six immune dysregulation (ID), two innate immune defects, and one in the autoinflammatory class. Overall, 23.5% of cases died, with disproportionate fatality rates among different IEI categories. PAD group had a relatively favorable outcome at any age, but CIDs and IDs were particularly vulnerable. At admission, presence of dyspnea was an independent risk for COVID-related death (OR: 2.630, 95% CI; 1.198-5.776, p < .001). Concerning predictive roles of laboratory markers at admission, deceased subjects compared to survived had significantly higher CRP, procalcitonin, Troponin-T, ferritin, and total-lung-score (p = .020, p = .003, p = .014, p = .013, p = .020; respectively), and lower absolute lymphocyte count, albumin, and trough IgG (p = .012, p = .022, p = .011; respectively).
Conclusion: Our data disclose a highly vulnerable IEI subgroup particularly disadvantaged for COVID-19 despite their youth. Future studies should address this vulnerability and consider giving priority to these subjects in SARS-Cov-2 therapy trials.
Keywords: COVID-19; SARS-Cov-2; inborn errors of immunity; outcome.
© 2021 European Academy of Allergy and Clinical Immunology and John Wiley & Sons Ltd.
Conflict of interest statement
All authors declare that they have no conflict of interest related to this work.
Figures



References
-
- WHO Coronavirus disease (COVID‐19) pandemic. 2020; https://www.WHO.int/emergencies/diseases/novel‐coronavirus‐2019, 2020. Accessed September 03, 2020.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

