The origins and potential future of SARS-CoV-2 variants of concern in the evolving COVID-19 pandemic
- PMID: 34314723
- PMCID: PMC8220957
- DOI: 10.1016/j.cub.2021.06.049
The origins and potential future of SARS-CoV-2 variants of concern in the evolving COVID-19 pandemic
Abstract
One year into the global COVID-19 pandemic, the focus of attention has shifted to the emergence and spread of SARS-CoV-2 variants of concern (VOCs). After nearly a year of the pandemic with little evolutionary change affecting human health, several variants have now been shown to have substantial detrimental effects on transmission and severity of the virus. Public health officials, medical practitioners, scientists, and the broader community have since been scrambling to understand what these variants mean for diagnosis, treatment, and the control of the pandemic through nonpharmaceutical interventions and vaccines. Here we explore the evolutionary processes that are involved in the emergence of new variants, what we can expect in terms of the future emergence of VOCs, and what we can do to minimise their impact.
Crown Copyright © 2021. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures
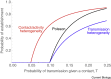


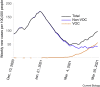


References
-
- Public Health England . 2020. Investigation of novel SARS-CoV-2 variant: Variant of Concern 202012/01. Version 1, release date 21/12/2020.https://www.gov.uk/government/publications/investigation-of-novel-sars-c...
-
- Volz E., Mishra S., Chand M., Barrett J.C., Johnson R., Geidelberg L., Hinsley W.R., Laydon D.J., Dabrera G., O'Toole A., et al. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature. 2021;593:266–269. - PubMed
-
- Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Msomi N., et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021;592:438–443. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

