N6-methyladenosine methyltransferases: functions, regulation, and clinical potential
- PMID: 34315512
- PMCID: PMC8313886
- DOI: 10.1186/s13045-021-01129-8
N6-methyladenosine methyltransferases: functions, regulation, and clinical potential
Abstract
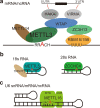
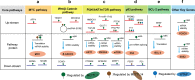
N6-methyladenosine (m6A) has emerged as an abundant modification throughout the transcriptome with widespread functions in protein-coding and noncoding RNAs. It affects the fates of modified RNAs, including their stability, splicing, and/or translation, and thus plays important roles in posttranscriptional regulation. To date, m6A methyltransferases have been reported to execute m6A deposition on distinct RNAs by their own or forming different complexes with additional partner proteins. In this review, we summarize the function of these m6A methyltransferases or complexes in regulating the key genes and pathways of cancer biology. We also highlight the progress in the use of m6A methyltransferases in mediating therapy resistance, including chemotherapy, targeted therapy, immunotherapy and radiotherapy. Finally, we discuss the current approaches and clinical potential of m6A methyltransferase-targeting strategies.
Keywords: Cancer; Drug discovery; Therapy resistance; m6A; m6A methyltransferase.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Functions of N6-methyladenosine and its role in cancer.Mol Cancer. 2019 Dec 4;18(1):176. doi: 10.1186/s12943-019-1109-9. Mol Cancer. 2019. PMID: 31801551 Free PMC article. Review.
-
The interplay between m6A RNA methylation and noncoding RNA in cancer.J Hematol Oncol. 2019 Nov 22;12(1):121. doi: 10.1186/s13045-019-0805-7. J Hematol Oncol. 2019. PMID: 31757221 Free PMC article. Review.
-
N6-methyladenosine modification in mRNA: machinery, function and implications for health and diseases.FEBS J. 2016 May;283(9):1607-30. doi: 10.1111/febs.13614. Epub 2015 Dec 31. FEBS J. 2016. PMID: 26645578 Review.
-
N6-Methyladenosine Methyltransferase Component KIAA1429 Is a Potential Target of Cancer Therapy.Biomolecules. 2024 Oct 17;14(10):1319. doi: 10.3390/biom14101319. Biomolecules. 2024. PMID: 39456252 Free PMC article. Review.
-
Research advances of N6-methyladenosine in diagnosis and therapy of pancreatic cancer.J Clin Lab Anal. 2022 Sep;36(9):e24611. doi: 10.1002/jcla.24611. Epub 2022 Jul 15. J Clin Lab Anal. 2022. PMID: 35837987 Free PMC article. Review.
Cited by
-
The Emerging Role of N6-Methyladenosine RNA Methylation as Regulators in Cancer Therapy and Drug Resistance.Front Pharmacol. 2022 Apr 6;13:873030. doi: 10.3389/fphar.2022.873030. eCollection 2022. Front Pharmacol. 2022. PMID: 35462896 Free PMC article. Review.
-
Dysfunctional epigenetic protein-coding gene-related signature is associated with the prognosis of pancreatic cancer based on histone modification and transcriptome analysis.Sci Rep. 2023 Jan 4;13(1):146. doi: 10.1038/s41598-022-27316-2. Sci Rep. 2023. PMID: 36599884 Free PMC article.
-
Role of m6A modification in regulating the PI3K/AKT signaling pathway in cancer.J Transl Med. 2023 Nov 1;21(1):774. doi: 10.1186/s12967-023-04651-0. J Transl Med. 2023. PMID: 37915034 Free PMC article. Review.
-
Decoding the regulatory roles of circular RNAs in cardiac fibrosis.Noncoding RNA Res. 2024 Dec 9;11:115-130. doi: 10.1016/j.ncrna.2024.11.007. eCollection 2025 Apr. Noncoding RNA Res. 2024. PMID: 39759175 Free PMC article. Review.
-
Von Hippel Lindau tumor suppressor controls m6A-dependent gene expression in renal tumorigenesis.J Clin Invest. 2024 Apr 15;134(8):e175703. doi: 10.1172/JCI175703. J Clin Invest. 2024. PMID: 38618952 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

