WIND (Workflow for pIRNAs aNd beyonD): a strategy for in-depth analysis of small RNA-seq data
- PMID: 34316353
- PMCID: PMC8276195
- DOI: 10.12688/f1000research.27868.3
WIND (Workflow for pIRNAs aNd beyonD): a strategy for in-depth analysis of small RNA-seq data
Abstract
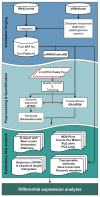
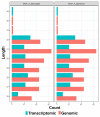
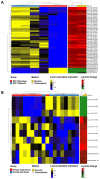
Current bioinformatics workflows for PIWI-interacting RNA (piRNA) analysis focus primarily on germline-derived piRNAs and piRNA-clusters. Frequently, they suffer from outdated piRNA databases, questionable quantification methods, and lack of reproducibility. Often, pipelines specific to miRNA analysis are used for the piRNA research in silico. Furthermore, the absence of a well-established database for piRNA annotation, as for miRNA, leads to uniformity issues between studies and generates confusion for data analysts and biologists. For these reasons, we have developed WIND ( Workflow for p IRNAs a Nd beyon D), a bioinformatics workflow that addresses the crucial issue of piRNA annotation, thereby allowing a reliable analysis of small RNA sequencing data for the identification of piRNAs and other small non-coding RNAs (sncRNAs) that in the past have been incorrectly classified as piRNAs. WIND allows the creation of a comprehensive annotation track of sncRNAs combining information available in RNAcentral, with piRNA sequences from piRNABank, the first database dedicated to piRNA annotation. WIND was built with Docker containers for reproducibility and integrates widely used bioinformatics tools for sequence alignment and quantification. In addition, it includes Bioconductor packages for exploratory data and differential expression analysis. Moreover, WIND implements a "dual" approach for the evaluation of sncRNAs expression level quantifying the aligned reads to the annotated genome and carrying out an alignment-free transcript quantification using reads mapped to the transcriptome. Therefore, a broader range of piRNAs can be annotated, improving their quantification and easing the subsequent downstream analysis. WIND performance has been tested with several small RNA-seq datasets, demonstrating how our approach can be a useful and comprehensive resource to analyse piRNAs and other classes of sncRNAs.
Keywords: ncRNA-expression; piRNA; small RNA sequencing; workflow.
Copyright: © 2021 Geles K et al.
Conflict of interest statement
No competing interests were disclosed.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

