Replication protein A: a multifunctional protein with roles in DNA replication, repair and beyond
- PMID: 34316690
- PMCID: PMC8210275
- DOI: 10.1093/narcan/zcaa022
Replication protein A: a multifunctional protein with roles in DNA replication, repair and beyond
Abstract
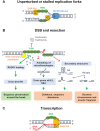
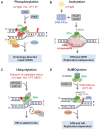
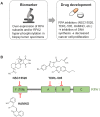
Single-stranded DNA (ssDNA) forms continuously during DNA replication and is an important intermediate during recombination-mediated repair of damaged DNA. Replication protein A (RPA) is the major eukaryotic ssDNA-binding protein. As such, RPA protects the transiently formed ssDNA from nucleolytic degradation and serves as a physical platform for the recruitment of DNA damage response factors. Prominent and well-studied RPA-interacting partners are the tumor suppressor protein p53, the RAD51 recombinase and the ATR-interacting proteins ATRIP and ETAA1. RPA interactions are also documented with the helicases BLM, WRN and SMARCAL1/HARP, as well as the nucleotide excision repair proteins XPA, XPG and XPF-ERCC1. Besides its well-studied roles in DNA replication (restart) and repair, accumulating evidence shows that RPA is engaged in DNA activities in a broader biological context, including nucleosome assembly on nascent chromatin, regulation of gene expression, telomere maintenance and numerous other aspects of nucleic acid metabolism. In addition, novel RPA inhibitors show promising effects in cancer treatment, as single agents or in combination with chemotherapeutics. Since the biochemical properties of RPA and its roles in DNA repair have been extensively reviewed, here we focus on recent discoveries describing several non-canonical functions.
© The Author(s) 2020. Published by Oxford University Press on behalf of NAR Cancer.
Figures

References
-
- Brill S.J., Stillman B. Yeast replication factor-A functions in the unwinding of the SV40 origin of DNA replication. Nature. 1989; 342:92–95. - PubMed
-
- Arunkumar A.I., Stauffer M.E., Bochkareva E., Bochkarev A., Chazin W.J. Independent and coordinated functions of replication protein A tandem high affinity single-stranded DNA binding domains. J. Biol. Chem. 2003; 278:41077–41082. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

