Molecular Characteristics of IS 1216 Carrying Multidrug Resistance Gene Cluster in Serotype III/Sequence Type 19 Group B Streptococcus
- PMID: 34319128
- PMCID: PMC8386385
- DOI: 10.1128/mSphere.00543-21
Molecular Characteristics of IS 1216 Carrying Multidrug Resistance Gene Cluster in Serotype III/Sequence Type 19 Group B Streptococcus
Abstract
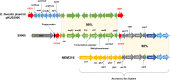
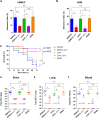
Streptococcus agalactiae is the leading cause of meningitis in newborns and a significant cause of invasive diseases in pregnant women and adults with underlying diseases. Antibiotic resistance against erythromycin and clindamycin in group B streptococcus (GBS) isolates has been increasing worldwide. GBS expresses the Srr1 and Srr2 proteins, which have important roles in bacterial infection. They have been investigated as novel vaccine candidates against GBS infection, with promising results. But a recent study detected non-srr1/2-expressing clinical isolates belonging to serotype III. Thus, we aimed to analyze the genotypes of non-srr1/2 GBS clinical isolates collected between 2013 and 2016 in South Korea. Forty-one (13.4%) of the 305 serotype III isolates were identified as non-srr1/2 strains, including sequence type 19 (ST19) (n = 16) and ST27 (n = 18) strains. The results of the comparative genomic analysis of the ST19/serotype III/non-srr1/2 strains further revealed four unique gene clusters. Site 4 in the srr1 gene locus was replaced by an lsa(E)-lnu(B)-aadK-aac-aph-aadE-carrying multidrug-resistant gene cluster flanked by two IS1216 transposases with 99% homology to the enterococcal plasmid pKUB3007-1. Despite the Srr1 and Srr2 deficiencies, which resulted in reduced fibrinogen binding, the adherence of non-srr1/2 strains to endothelial and epithelial cells was comparable to that of Srr1- or Srr2-expressing strains. Moreover, their virulence in mouse models of meningitis was not significantly affected. Furthermore, additional adhesin-encoding genes, including a gene encoding a BspA-like protein, which may contribute to colonization by non-srr1/2 strains, were identified via whole-genome analysis. Thus, our study provides important findings that can aid in the development of vaccines and antibiotics against GBS. IMPORTANCE Most previously isolated group B streptococcus (GBS) strains express either the Srr1 or Srr2 glycoprotein, which plays an important role in bacterial colonization and invasion. These glycoproteins are potential protein vaccine candidates. In this study, we first report GBS clinical isolates in which the srr1/2 gene was deleted or replaced with foreign genes. Despite Srr1/2 deficiency, in vitro adherence to mammalian cells and in vivo virulence in murine models were not affected, suggesting that the isolates might have another adherence mechanism that enhanced their virulence aside from Srr1/2-fibrinogen-mediated adherence. In addition, several non-srr1/2 isolates replaced the srr1/2 gene with the lnu(B) and lsa(E) antibiotic resistance genes flanked by IS1216, effectively causing multidrug resistance. Collectively, we believe that our study identifies the underlying genes responsible for the pathogenesis of new GBS serotype III. Furthermore, our study emphasizes the need for alternative antibiotics for patients who are allergic to β-lactams and for those who are pregnant.
Keywords: IS1216; ST19; Streptococcus agalactiae; multidrug resistance gene; srr1/2.
Figures


Similar articles
-
Genomic insights into the diversity, virulence, and antimicrobial resistance of group B Streptococcus clinical isolates from Saudi Arabia.Front Cell Infect Microbiol. 2024 Apr 22;14:1377993. doi: 10.3389/fcimb.2024.1377993. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38711928 Free PMC article.
-
New genetic context of lnu(B) composed of two multi-resistance gene clusters in clinical Streptococcus agalactiae ST-19 strains.Antimicrob Resist Infect Control. 2019 Jul 15;8:117. doi: 10.1186/s13756-019-0563-x. eCollection 2019. Antimicrob Resist Infect Control. 2019. PMID: 31346458 Free PMC article.
-
Predomination of hypervirulent ST283 and genetic diversity of levofloxacin resistance in multidrug-resistant, hypervirulent Streptococcus agalactiae in Thailand.J Med Microbiol. 2025 Mar;74(3):001970. doi: 10.1099/jmm.0.001970. J Med Microbiol. 2025. PMID: 40100264 Free PMC article.
-
Streptococcus agalactiae maternal colonization, antibiotic resistance and serotype profiles in Africa: a meta-analysis.Ann Clin Microbiol Antimicrob. 2019 Mar 28;18(1):14. doi: 10.1186/s12941-019-0313-1. Ann Clin Microbiol Antimicrob. 2019. PMID: 30922308 Free PMC article.
-
Group B streptococcal haemolysin and pigment, a tale of twins.FEMS Microbiol Rev. 2014 Sep;38(5):932-46. doi: 10.1111/1574-6976.12071. Epub 2014 Apr 4. FEMS Microbiol Rev. 2014. PMID: 24617549 Free PMC article. Review.
Cited by
-
Emergence of High-Level Gentamicin Resistance in Streptococcus agalactiae Hypervirulent Serotype IV ST1010 (CC452) Strains by Acquisition of a Novel Integrative and Conjugative Element.Antibiotics (Basel). 2024 May 26;13(6):491. doi: 10.3390/antibiotics13060491. Antibiotics (Basel). 2024. PMID: 38927158 Free PMC article.
-
A Survey of Chinese Pig Farms and Human Healthcare Isolates Reveals Separate Human and Animal Methicillin-Resistant Staphylococcus aureus Populations.Adv Sci (Weinh). 2022 Feb;9(4):e2103388. doi: 10.1002/advs.202103388. Epub 2021 Dec 11. Adv Sci (Weinh). 2022. PMID: 34894204 Free PMC article.
-
Genomic characterization of group B Streptococcus from Argentina: insights into prophage diversity, virulence factors and antibiotic resistance genes.Microb Genom. 2025 Apr;11(4):001399. doi: 10.1099/mgen.0.001399. Microb Genom. 2025. PMID: 40266661 Free PMC article.
-
Recent development and fighting strategies for lincosamide antibiotic resistance.Clin Microbiol Rev. 2024 Jun 13;37(2):e0016123. doi: 10.1128/cmr.00161-23. Epub 2024 Apr 18. Clin Microbiol Rev. 2024. PMID: 38634634 Free PMC article. Review.
-
Long-Term Co-Circulation of Host-Specialist and Host-Generalist Lineages of Group B Streptococcus in Brazilian Dairy Cattle with Heterogeneous Antimicrobial Resistance Profiles.Antibiotics (Basel). 2024 Apr 25;13(5):389. doi: 10.3390/antibiotics13050389. Antibiotics (Basel). 2024. PMID: 38786118 Free PMC article.
References
-
- Falagas ME, Rosmarakis ES, Avramopoulos I, Vakalis N. 2006. Streptococcus agalactiae infections in non-pregnant adults: single center experience of a growing clinical problem. Med Sci Monit 12:CR447–CR451. - PubMed
-
- Centers for Disease Control and Prevention. 2016. Group B Streptococcus (GBS) 2016. Centers for Disease Control and Prevention, Atlanta, GA. https://www.cdc.gov/groupbstrep/index.html. Accessed 1 November 2017.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

