Development of a Biocontained Toluene-Degrading Bacterium for Environmental Protection
- PMID: 34319138
- PMCID: PMC8552602
- DOI: 10.1128/Spectrum.00259-21
Development of a Biocontained Toluene-Degrading Bacterium for Environmental Protection
Abstract
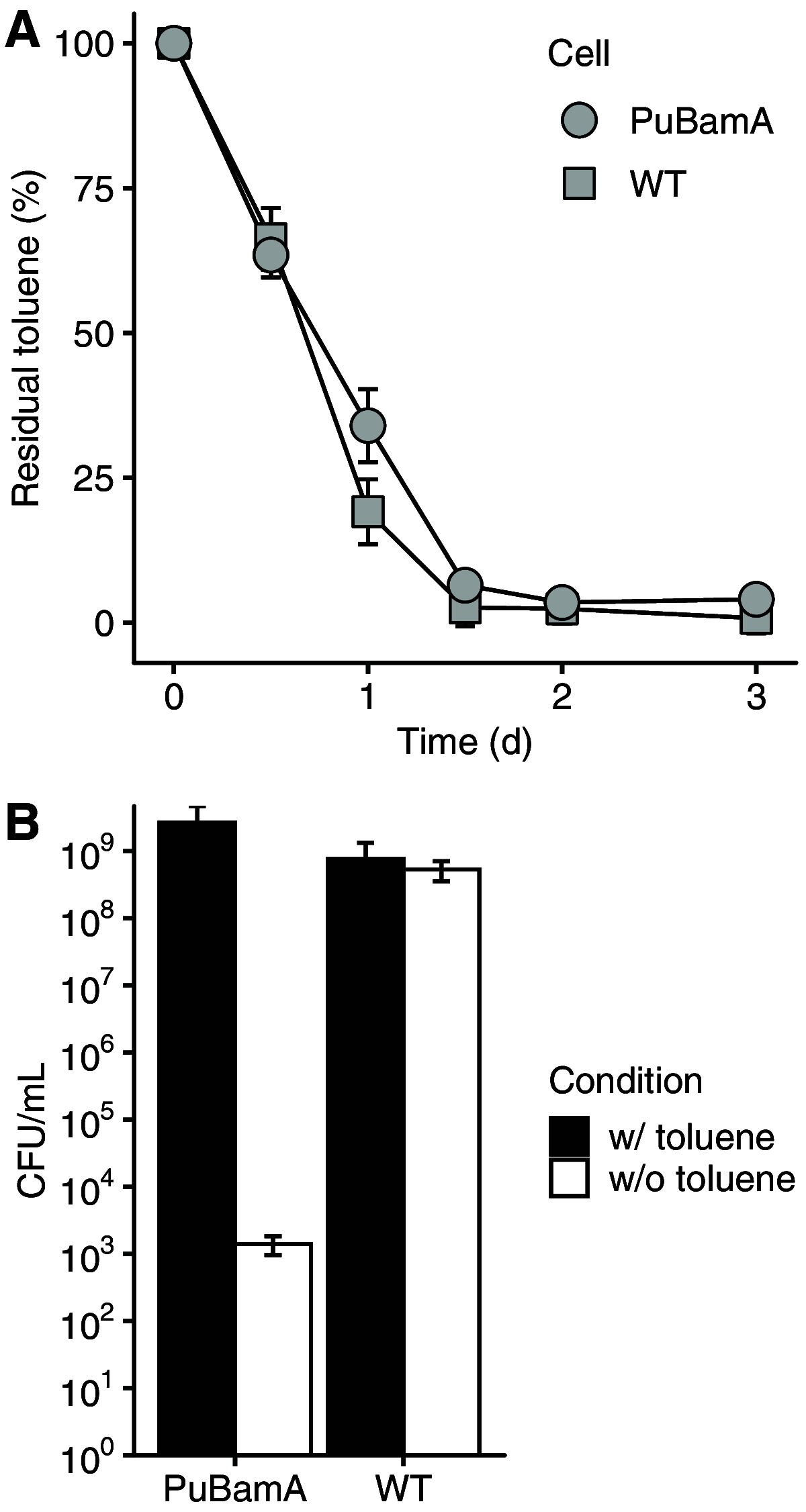
Biocontainment is a safeguard strategy for preventing uncontrolled proliferation of genetically engineered microorganisms (GEMs) in the environment. Biocontained GEMs are designed to survive only in the presence of a specific molecule. The design of a pollutant-degrading and pollutant-dependent GEM prevents its proliferation after cleaning the environment. In this study, we present a biocontained toluene-degrading bacterium based on Acinetobacter sp. Tol 5. The bamA gene, which encodes an essential outer membrane protein, was deleted from the chromosome of Tol 5 but complemented with a plasmid carrying a bamA gene regulated by the Pu promoter and the regulatory protein XylR. The resultant strain (PuBamA) degraded toluene, similarly to the wild-type Tol 5. Although the cell growth of the PuBamA strain was remarkably inhibited after toluene depletion, escape mutants emerged at a frequency of 1 per 5.3 × 10-7 cells. Analyses of escape mutants revealed that insertion sequences (ISs) carrying promoters were inserted between the Pu promoter and the bamA gene on the complemented plasmid. MinION deep sequencing of the plasmids extracted from the escape mutants enabled the identification of three types of ISs involved in the emergence of escape mutants, suggesting a strategy for reducing it. IMPORTANCE GEMs are beneficial for various applications, including environmental protection. However, the risks of GEM release into the environment have been debated for a long time. If a pollutant is employed as a specific molecule for a biocontainment system, GEMs capable of degrading pollutants are available for environmental protection. Nevertheless, to our knowledge, biocontained degraders for real pollutants have not been reported in academic journals so far. This is possibly due to the difficulty in the expression of enzymes for degrading pollutants in a tractable bacterium such as Escherichia coli. On the other hand, bacteria with enzymes for degrading pollutants are often intractable as a host of GEMs due to the shortage of tools for genetic manipulation. This study reports the feasibility of a biocontainment strategy for a toluene degrader. Our results provide useful insights into the construction of a GEM biocontainment system for environmental protection.
Keywords: MinION; biocontainment; insertion sequence; synthetic biology.
Figures




Similar articles
-
Metabolic engineering of bacteria for environmental applications: construction of Pseudomonas strains for biodegradation of 2-chlorotoluene.J Biotechnol. 2001 Feb 13;85(2):103-13. doi: 10.1016/s0168-1656(00)00367-9. J Biotechnol. 2001. PMID: 11165359
-
The impact of succinate trace on pWW0 and ortho-cleavage pathway transcription in Pseudomonas putida mt-2 during toluene biodegradation.Bioresour Technol. 2017 Jun;234:397-405. doi: 10.1016/j.biortech.2017.03.082. Epub 2017 Mar 16. Bioresour Technol. 2017. PMID: 28347959
-
Transcriptional wiring of the TOL plasmid regulatory network to its host involves the submission of the sigma54-promoter Pu to the response regulator PprA.Mol Microbiol. 2008 Aug;69(3):698-713. doi: 10.1111/j.1365-2958.2008.06321.x. Mol Microbiol. 2008. PMID: 19138193
-
Be a GEM: Biocontained, environmentally applied, genetically engineered microbes.Adv Drug Deliv Rev. 2025 Jun;221:115578. doi: 10.1016/j.addr.2025.115578. Epub 2025 Apr 11. Adv Drug Deliv Rev. 2025. PMID: 40222715 Review.
-
Fate of genetically-engineered bacteria in activated sludge microcosms.Schriftenr Ver Wasser Boden Lufthyg. 1988;78:267-76. Schriftenr Ver Wasser Boden Lufthyg. 1988. PMID: 3074483 Review.
Cited by
-
Genetically stable kill-switch using "demon and angel" expression construct of essential genes.Front Bioeng Biotechnol. 2024 Feb 28;12:1365870. doi: 10.3389/fbioe.2024.1365870. eCollection 2024. Front Bioeng Biotechnol. 2024. PMID: 38481573 Free PMC article.
-
The elimination of two restriction enzyme genes allows for electroporation-based transformation and CRISPR-Cas9-based base editing in the non-competent Gram-negative bacterium Acinetobacter sp. Tol 5.Appl Environ Microbiol. 2024 Jun 18;90(6):e0040024. doi: 10.1128/aem.00400-24. Epub 2024 May 9. Appl Environ Microbiol. 2024. PMID: 38722179 Free PMC article.
-
The Japan-UK Synthetic Biology Conference, Spring 2025: Strengthening Global Links to Engineer Biology.ACS Synth Biol. 2025 Jun 20;14(6):1873-1878. doi: 10.1021/acssynbio.5c00232. ACS Synth Biol. 2025. PMID: 40538270 Free PMC article.
-
Prolonging genetic circuit stability through adaptive evolution of overlapping genes.Nucleic Acids Res. 2023 Jul 21;51(13):7094-7108. doi: 10.1093/nar/gkad484. Nucleic Acids Res. 2023. PMID: 37260076 Free PMC article.
-
Stability, robustness, and containment: preparing synthetic biology for real-world deployment.Curr Opin Biotechnol. 2023 Feb;79:102880. doi: 10.1016/j.copbio.2022.102880. Epub 2023 Jan 6. Curr Opin Biotechnol. 2023. PMID: 36621221 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

