IFITM Proteins That Restrict the Early Stages of Respiratory Virus Infection Do Not Influence Late-Stage Replication
- PMID: 34319159
- PMCID: PMC8475524
- DOI: 10.1128/JVI.00837-21
IFITM Proteins That Restrict the Early Stages of Respiratory Virus Infection Do Not Influence Late-Stage Replication
Abstract
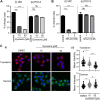
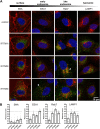
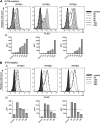
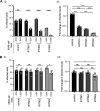
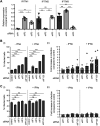
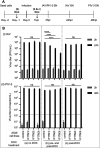
Interferon-induced transmembrane (IFITM) proteins inhibit a broad range of enveloped viruses by blocking entry into host cells. We used an inducible overexpression system to investigate if IFITM1, IFITM2, and IFITM3 could modulate early and/or late stages of influenza A virus (IAV) or parainfluenza virus 3 (PIV-3) infection in human A549 airway epithelial cells. IAV and PIV-3 represent respiratory viruses which utilize distinct cellular entry pathways. We verify entry by endocytosis for IAV, whereas PIV-3 infection was consistent with fusion at the plasma membrane. Following induction prior to infection, all three IFITM proteins restricted the percentage of IAV-infected cells at 8 hours postinfection. In contrast, prior induction of IFITM1 and IFITM2 did not inhibit PIV-3 infection, although a modest reduction was observed with IFITM3. Small interfering RNA (siRNA)-mediated knockdown of endogenous IFITM1, IFITM2, and IFITM3 expression, in the presence or absence of pretreatment with type I interferon, resulted in increased IAV, but not PIV-3, infection. This finding suggests that while all three IFITMs display antiviral activity against IAV, they do not restrict the early stages of PIV-3 infection. IAV and PIV-3 infection culminates in viral egress through budding at the plasma membrane. Inducible expression of IFITM1, IFITM2, or IFITM3 immediately after infection did not impact titers of infectious virus released from IAV- or PIV-3-infected cells. Our findings show that IFITM proteins differentially restrict the early stages of infection of two respiratory viruses with distinct cellular entry pathways but do not influence the late stages of replication for either virus. IMPORTANCE Interferon-induced transmembrane (IFITM) proteins restrict the initial stages of infection for several respiratory viruses; however, their potential to modulate the later stages of virus replication has not been explored. In this study, we highlight the utility of an inducible overexpression system to assess the impact of IFITM proteins on either early- or late-stage replication of two respiratory viruses. We demonstrate antiviral activity by IFITM1, IFITM2, and IFITM3 against influenza A virus (IAV) but not parainfluenza virus 3 (PIV-3) during the early stages of cellular infection. Furthermore, IFITM induction following IAV or PIV-3 infection does not restrict the late stages of replication of either virus. Our findings show that IFITM proteins can differentially restrict the early stages of infection of two viruses with distinct cellular entry pathways and yet do not influence the late stages of replication for either virus.
Keywords: IFITM; host restriction factors; influenza; parainfluenza virus.
Figures

Similar articles
-
Duck Interferon-Inducible Transmembrane Protein 3 Mediates Restriction of Influenza Viruses.J Virol. 2015 Oct 14;90(1):103-16. doi: 10.1128/JVI.01593-15. Print 2016 Jan 1. J Virol. 2015. PMID: 26468537 Free PMC article.
-
Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus.PLoS Pathog. 2011 Jan 6;7(1):e1001258. doi: 10.1371/journal.ppat.1001258. PLoS Pathog. 2011. PMID: 21253575 Free PMC article.
-
pH Optimum of Hemagglutinin-Mediated Membrane Fusion Determines Sensitivity of Influenza A Viruses to the Interferon-Induced Antiviral State and IFITMs.J Virol. 2017 May 12;91(11):e00246-17. doi: 10.1128/JVI.00246-17. Print 2017 Jun 1. J Virol. 2017. PMID: 28356532 Free PMC article.
-
IFITM proteins: Understanding their diverse roles in viral infection, cancer, and immunity.J Biol Chem. 2023 Jan;299(1):102741. doi: 10.1016/j.jbc.2022.102741. Epub 2022 Nov 23. J Biol Chem. 2023. PMID: 36435199 Free PMC article. Review.
-
Discovery and Characterization of IFITM S-Palmitoylation.Viruses. 2023 Nov 28;15(12):2329. doi: 10.3390/v15122329. Viruses. 2023. PMID: 38140570 Free PMC article. Review.
Cited by
-
Differential Transcriptomics Analysis of IPEC-J2 Cells Single or Coinfected With Porcine Epidemic Diarrhea Virus and Transmissible Gastroenteritis Virus.Front Immunol. 2022 Mar 25;13:844657. doi: 10.3389/fimmu.2022.844657. eCollection 2022. Front Immunol. 2022. PMID: 35401515 Free PMC article.
-
Differential Leukocyte Expression of IFITM1 and IFITM3 in Patients with Severe Pandemic Influenza A(H1N1) and COVID-19.J Interferon Cytokine Res. 2022 Aug;42(8):430-443. doi: 10.1089/jir.2022.0036. Epub 2022 Jun 14. J Interferon Cytokine Res. 2022. PMID: 35708622 Free PMC article.
-
Positive Regulation of the Antiviral Activity of Interferon-Induced Transmembrane Protein 3 by S-Palmitoylation.Front Immunol. 2022 Jun 13;13:919477. doi: 10.3389/fimmu.2022.919477. eCollection 2022. Front Immunol. 2022. PMID: 35769480 Free PMC article. Review.
-
Potential antiviral effects of the marine probiotic Paraliobacillus zengyii on the respiratory syncytial virus.mLife. 2025 Jun 18;4(3):249-258. doi: 10.1002/mlf2.70015. eCollection 2025 Jun. mLife. 2025. PMID: 40600055 Free PMC article.
-
Progress in Pseudotyping Lentiviral Vectors Towards Cell-Specific Gene Delivery In Vivo.Viruses. 2025 May 31;17(6):802. doi: 10.3390/v17060802. Viruses. 2025. PMID: 40573393 Free PMC article. Review.
References
-
- Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, Adams DJ, Xavier RJ, Farzan M, Elledge SJ. 2009. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139:1243–1254. 10.1016/j.cell.2009.12.017. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- APP1143154/Department of Health, Australian Government | National Health and Medical Research Council (NHMRC)
- APP1083307/Department of Health, Australian Government | National Health and Medical Research Council (NHMRC)
- APP1184532/Department of Health, Australian Government | National Health and Medical Research Council (NHMRC)
LinkOut - more resources
Full Text Sources
Medical

