Synthesis of Oligonucleotides Containing Trans Mitomycin C DNA Adducts at N6 of Adenine and N2 of Guanine
- PMID: 34319608
- PMCID: PMC8516704
- DOI: 10.1002/chem.202102338
Synthesis of Oligonucleotides Containing Trans Mitomycin C DNA Adducts at N6 of Adenine and N2 of Guanine
Abstract
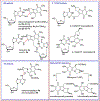
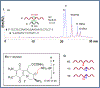
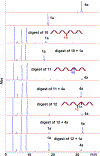
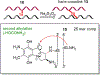
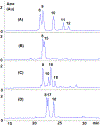
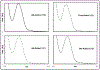
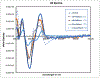
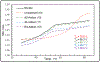
Mitomycin C, (MC), an antitumor drug, is a DNA alkylating agent currently used in the clinics. Inert in its native form, MC is reduced to reactive mitosenes, which undergo nucleophilic attack by guanine or adenine bases in DNA to form monoadducts as well as interstrand crosslinks (ICLs). Although ICLs are considered the most cytotoxic lesions, the role of each individual adduct in the drug's cytotoxicity is still not fully understood. Synthetic routes have been developed to access modified oligonucleotides containing dG MC-monoadducts and dG-MC-dG ICL at a single position of their base sequences to investigate the biological effects of these adducts. However, until now, oligonucleotides containing monoadducts formed by MC at the adenine base had not been available, thus preventing the examination of the role played by these lesions in the toxicity of MC. Here, we present a route to access these substrates. Structural proof of the adducted oligonucleotides were provided by enzymatic digestion to nucleosides and high-resolution mass spectral analysis. Additionally, parent oligonucleotides containing a dG monoadduct and a dG-MC-dG ICL were also produced. The stability and physical properties of all substrates were compared via CD spectroscopy and UV melting temperature studies. Finally, virtual models were created to explore the conformational space and structural features of these MC-DNA complexes.
Keywords: alkylation; conformational studies; deoxyadenosine adducts; interstrand crosslinks; site-specific mitomycin C.
© 2021 Wiley-VCH GmbH.
Figures



Similar articles
-
Synthesis of Oligonucleotides containing the cis-Interstrand Crosslink Produced by Mitomycins in their Reaction with DNA.Chemistry. 2020 Oct 1;26(55):12570-12578. doi: 10.1002/chem.202002452. Epub 2020 Sep 3. Chemistry. 2020. PMID: 32574396 Free PMC article.
-
Insight Into Factors Governing Formation, Synthesis and Stereochemical Configuration of DNA Adducts Formed by Mitomycins.Chem Rec. 2023 Jan;23(1):e202200193. doi: 10.1002/tcr.202200193. Epub 2022 Oct 17. Chem Rec. 2023. PMID: 36251922 Review.
-
Relative toxicities of DNA cross-links and monoadducts: new insights from studies of decarbamoyl mitomycin C and mitomycin C.Chem Res Toxicol. 2002 Nov;15(11):1398-406. doi: 10.1021/tx020044g. Chem Res Toxicol. 2002. PMID: 12437330
-
Synthesis of Mitomycin C and decarbamoylmitomycin C N6 deoxyadenosine-adducts.Bioorg Chem. 2019 Nov;92:103280. doi: 10.1016/j.bioorg.2019.103280. Epub 2019 Sep 12. Bioorg Chem. 2019. PMID: 31539740 Free PMC article.
-
A mitomycin-N6-deoxyadenosine adduct isolated from DNA.Chem Res Toxicol. 1998 Mar;11(3):203-10. doi: 10.1021/tx970205u. Chem Res Toxicol. 1998. PMID: 9544618
Cited by
-
Mitomycin C and its analog trigger cytotoxicity in MCF-7 and K562 cancer cells through the regulation of RAS and MAPK/ERK pathways.Chem Biol Interact. 2024 May 25;395:111007. doi: 10.1016/j.cbi.2024.111007. Epub 2024 Apr 18. Chem Biol Interact. 2024. PMID: 38642817 Free PMC article.
-
Stereoisomeric mitomycins interstrand crosslinks differently impact gene expression in MCF-7 and K562 cancer cells.Chem Biol Interact. 2025 Aug 25;417:111564. doi: 10.1016/j.cbi.2025.111564. Epub 2025 May 16. Chem Biol Interact. 2025. PMID: 40383468
References
-
- Hata T, Hoshi T, Kanamori K, Matsumae A, Sano Y, Shima T, Sugawara R, J. Antibiot 1956, 9, 141–146. - PubMed
-
- Bradner WT, Cancer Treat. Rev 2001, 27, 35–50. - PubMed
-
- Verweij J, Pinedo H, in Cancer Chemotherapy and Biological Modifiers: Annual 11 (Eds.: Pinedo HM, Chabner BA, Longo DL), Elsevier Science Publishers B.V., Amsterdam, 1990, p. 67.
-
- Chabner BA, Amrein PC, Druker BJ, Michaelson MD, Mitsiades CS, Goss PE, Ryan DP, Ramachandra S, Richardson PJ, Supko JG, Wilson WH, “Antineoplastic Agents” in Goodman & Gilman’s The Pharmacological Basis of Therapeutics (Eds.: Brunton LL, Lazo JS, Parker KL), McGraw-Hill Publishers, New York, 2005, pp. 1315–1403.
-
- Paz MM, Pritsos CA, “The Molecular Toxicology of Mitomycin C” in Advances Molecular Toxicology Vol. 6 (Ed.: Fishbein JC), Elsevier Science Publishers B.V., Amsterdam, 2012, pp. 244–286.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

