Phosphorylation of JIP4 at S730 Presents Antiviral Properties against Influenza A Virus Infection
- PMID: 34319782
- PMCID: PMC8475540
- DOI: 10.1128/JVI.00672-21
Phosphorylation of JIP4 at S730 Presents Antiviral Properties against Influenza A Virus Infection
Abstract
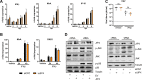
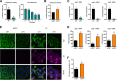
Influenza A virus (IAV) is the causative agent of flu disease that results in annual epidemics and occasional pandemics. IAV alters several signaling pathways of the cellular host response in order to promote its replication. Therefore, some of these pathways can serve as targets for novel antiviral agents. Here, we show that c-Jun NH2-terminal kinase (JNK)-interacting protein 4 (JIP4) is dynamically phosphorylated in IAV infection. The lack of JIP4 resulted in higher virus titers, with significant differences in viral protein and mRNA accumulation as early as within the first replication cycle. In accordance, decreased IAV titers and protein accumulation were observed during the overexpression of JIP4. Strikingly, the antiviral function of JIP4 does not originate from modulation of JNK or p38 mitogen-activated protein kinase (MAPK) pathways or from altered expression of interferons or interferon-stimulated genes but rather originates from a direct reduction of viral polymerase activity. Furthermore, the interference of JIP4 with IAV replication seems to be linked to the phosphorylation of the serine at position 730 that is sufficient to impede the viral polymerase. Collectively, we provide evidence that JIP4, a host protein modulated in IAV infection, exhibits antiviral properties that are dynamically controlled by its phosphorylation at S730. IMPORTANCE Influenza A virus (IAV) infection is a world health concern, and current treatment options encounter high rates of resistance. Our group investigates host pathways modified in IAV infection as promising new targets. The host protein JIP4 is dynamically phosphorylated in IAV infection. JIP4 absence resulted in higher virus titers and viral protein and mRNA accumulation within the first replication cycle. Accordingly, decreased IAV titers and protein accumulation were observed during JIP4 overexpression. Strikingly, the antiviral function of JIP4 does not originate from modulation of JNK or p38 MAPK pathways or from altered expression of interferons or interferon-stimulated genes but rather originates from a reduction in viral polymerase activity. The interference of JIP4 with IAV replication is linked to the phosphorylation of serine 730. We provide evidence that JIP4, a host protein modulated in IAV infection, exhibits antiviral properties that are dynamically controlled by its phosphorylation at S730.
Keywords: JIP4; host pathway; influenza virus; protein phosphorylation.
Figures



References
-
- Laure M, Hamza H, Koch-Heier J, Quernheim M, Müller C, Schreiber A, Müller G, Pleschka S, Ludwig S, Planz O. 2020. Antiviral efficacy against influenza virus and pharmacokinetic analysis of a novel MEK-inhibitor, ATR-002, in cell culture and in the mouse model. Antiviral Res 178:104806. 10.1016/j.antiviral.2020.104806. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

