nsP4 Is a Major Determinant of Alphavirus Replicase Activity and Template Selectivity
- PMID: 34319783
- PMCID: PMC8475546
- DOI: 10.1128/JVI.00355-21
nsP4 Is a Major Determinant of Alphavirus Replicase Activity and Template Selectivity
Abstract
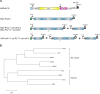
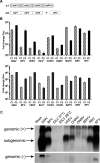
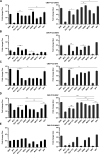
Alphaviruses have positive-strand RNA genomes containing two open reading frames (ORFs). The first ORF encodes the nonstructural (ns) polyproteins P123 and P1234 that act as precursors for the subunits of the viral RNA replicase (nsP1 to nsP4). Processing of P1234 leads to the formation of a negative-strand replicase consisting of nsP4 (RNA polymerase) and P123 components. Subsequent processing of P123 results in a positive-strand replicase. The second ORF encoding the structural proteins is expressed via the synthesis of a subgenomic RNA. Alphavirus replicase is capable of using template RNAs that contain essential cis-active sequences. Here, we demonstrate that the replicases of nine alphaviruses, expressed in the form of separate P123 and nsP4 components, are active. Their activity depends on the abundance of nsP4. The match of nsP4 to its template strongly influences efficient subgenomic RNA synthesis. nsP4 of Barmah Forest virus (BFV) formed a functional replicase only with matching P123, while nsP4s of other alphaviruses were compatible also with several heterologous P123s. The P123 components of Venezuelan equine encephalitis virus and Sindbis virus (SINV) required matching nsP4s, while P123 of other viruses could form active replicases with different nsP4s. Chimeras of Semliki Forest virus, harboring the nsP4 of chikungunya virus, Ross River virus, BFV, or SINV were viable. In contrast, chimeras of SINV, harboring an nsP4 from different alphaviruses, exhibited a temperature-sensitive phenotype. These findings highlight the possibility for formation of new alphaviruses via recombination events and provide a novel approach for the development of attenuated chimeric viruses for vaccination strategies. IMPORTANCE A key element of every virus with an RNA genome is the RNA replicase. Understanding the principles of RNA replicase formation and functioning is therefore crucial for understanding and responding to the emergence of new viruses. Reconstruction of the replicases of nine alphaviruses from nsP4 and P123 polyproteins revealed that the nsP4 of the majority of alphaviruses, including the mosquito-specific Eilat virus, could form a functional replicase with P123 originating from a different virus, and the corresponding chimeric viruses were replication-competent. nsP4 also had an evident role in determining the template RNA preference and the efficiency of RNA synthesis. The revealed broad picture of the compatibility of the replicase components of alphaviruses is important for understanding the formation and functioning of the alphavirus RNA replicase and highlights the possibilities for recombination between different alphavirus species.
Keywords: RNA polymerases; RNA replication; alphavirus; genetic recombination; replicase.
Figures
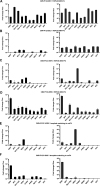

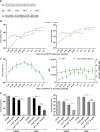
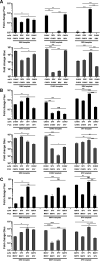
References
-
- Nasar F, Palacios G, Gorchakov RV, Guzman H, Da Rosa APT, Savji N, Popov VL, Sherman MB, Lipkin WI, Tesh RB, Weaver SC. 2012. Eilat virus, a unique alphavirus with host range restricted to insects by RNA replication. Proc Natl Acad Sci USA 109:14622–14627. 10.1073/pnas.1204787109. - DOI - PMC - PubMed
-
- Torii S, Orba Y, Hang’ombe BM, Mweene AS, Wada Y, Anindita PD, Phongphaew W, Qiu Y, Kajihara M, Mori-Kajihara A, Eto Y, Harima H, Sasaki M, Carr M, Hall WW, Eshita Y, Abe T, Sawa H. 2018. Discovery of Mwinilunga alphavirus: a novel alphavirus in Culex mosquitoes in Zambia. Virus Res 250:31–36. 10.1016/j.virusres.2018.04.005. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BBS/E/I/00007033 BBS/E/I/00007038 and BBS/E/I/00007039/UKRI | Biotechnology and Biological Sciences Research Council (BBSRC)
- MC_PC_16028/MRC_/Medical Research Council/United Kingdom
- PRG1154/Estonian Research Council
- 2014-2020.4.01.15-013/EC | European Regional Development Fund (ERDF)
- KAN1526318N/Research Foundation Flanders
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

