Emergence of equine-like G3 strains as the dominant rotavirus among children under five with diarrhea in Sabah, Malaysia during 2018-2019
- PMID: 34320003
- PMCID: PMC8318246
- DOI: 10.1371/journal.pone.0254784
Emergence of equine-like G3 strains as the dominant rotavirus among children under five with diarrhea in Sabah, Malaysia during 2018-2019
Abstract
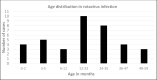
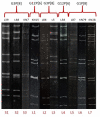
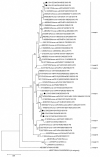
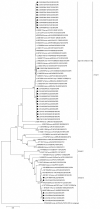
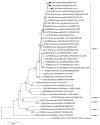
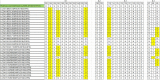
Rotavirus infection is a dilemma for developing countries, including Malaysia. Although commercial rotavirus vaccines are available, these are not included in Malaysia's national immunization program. A scarcity of data about rotavirus genotype distribution could be partially to blame for this policy decision, because there are no data for rotavirus genotype distribution in Malaysia over the past 20 years. From January 2018 to March 2019, we conducted a study to elucidate the rotavirus burden and genotype distribution in the Kota Kinabalu and Kunak districts of the state of Sabah. Stool specimens were collected from children under 5 years of age, and rotavirus antigen in these samples was detected using commercially available kit. Electropherotypes were determined by polyacrylamide gel electrophoresis of genomic RNA. G and P genotypes were determined by RT-PCR using type specific primers. The nucleotide sequence of the amplicons was determined by Sanger sequencing and phylogenetic analysis was performed by neighbor-joining method. Rotavirus was identified in 43 (15.1%) children with watery diarrhea. The male:female ratio (1.9:1) of the rotavirus-infected children clearly showed that it affected predominantly boys, and children 12-23 months of age. The genotypes identified were G3P[8] (74% n = 31), followed by G1P[8] (14% n = 6), G12P[6](7% n = 3), G8P[8](3% n = 1), and GxP[8] (3% n = 1). The predominant rotavirus circulating among the children was the equine-like G3P[8] (59.5% n = 25) with a short electropherotype. Eleven electropherotypes were identified among 34 strains, indicating substantial diversity among the circulating strains. The circulating genotypes were also phylogenetically diverse and related to strains from several different countries. The antigenic epitopes present on VP7 and VP4 of Sabahan G3 and equine-like G3 differed considerably from that of the RotaTeq vaccine strain. Our results also indicate that considerable genetic exchange is occurring in Sabahan strains. Sabah is home to a number of different ethnic groups, some of which culturally are in close contact with animals, which might contribute to the evolution of diverse rotavirus strains. Sabah is also a popular tourist destination, and a large number of tourists from different countries possibly contributes to the diversity of circulating rotavirus genotypes. Considering all these factors which are contributing rotavirus genotype diversity, continuous surveillance of rotavirus strains is of utmost importance to monitor the pre- and post-vaccination efficacy of rotavirus vaccines in Sabah.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Upsurge and spread of G3 rotaviruses in Eastern India (2014-2016): Full genome analyses reveals heterogeneity within Wa-like genomic constellation.Infect Genet Evol. 2018 Sep;63:158-174. doi: 10.1016/j.meegid.2018.05.026. Epub 2018 May 26. Infect Genet Evol. 2018. PMID: 29842980
-
Increase in rotavirus prevalence with the emergence of genotype G9P[8] in replacement of genotype G12P[6] in Sabah, Malaysia.Arch Virol. 2023 Jun 3;168(6):173. doi: 10.1007/s00705-023-05803-9. Arch Virol. 2023. PMID: 37269384 Free PMC article.
-
Diversity of rotavirus strains circulating in Northern Brazil after introduction of a rotavirus vaccine: high prevalence of G3P[6] genotype.J Med Virol. 2014 Jun;86(6):1065-72. doi: 10.1002/jmv.23797. Epub 2013 Oct 17. J Med Virol. 2014. PMID: 24136444
-
Rotavirus Strain Trends in United States, 2009-2016: Results from the National Rotavirus Strain Surveillance System (NRSSS).Viruses. 2022 Aug 15;14(8):1775. doi: 10.3390/v14081775. Viruses. 2022. PMID: 36016397 Free PMC article. Review.
-
Rotavirus infections and their genotype distribution pre- and post-vaccine introduction in Ethiopia: a systemic review and meta-analysis.BMC Infect Dis. 2024 Aug 16;24(1):836. doi: 10.1186/s12879-024-09754-7. BMC Infect Dis. 2024. PMID: 39152402 Free PMC article.
Cited by
-
Serological Humoral Immunity Following Natural Infection of Children with High Burden Gastrointestinal Viruses.Viruses. 2021 Oct 9;13(10):2033. doi: 10.3390/v13102033. Viruses. 2021. PMID: 34696463 Free PMC article. Review.
-
Phylodynamic characteristics of reassortant DS-1-like G3P[8]-strains of rotavirus type A isolated in Nizhny Novgorod (Russia).Braz J Microbiol. 2023 Dec;54(4):2867-2877. doi: 10.1007/s42770-023-01155-3. Epub 2023 Oct 28. Braz J Microbiol. 2023. PMID: 37897627 Free PMC article.
-
Acute Infectious Gastroenteritis: The Causative Agents, Omics-Based Detection of Antigens and Novel Biomarkers.Children (Basel). 2021 Dec 2;8(12):1112. doi: 10.3390/children8121112. Children (Basel). 2021. PMID: 34943308 Free PMC article. Review.
-
Emergence and dissemination of equine-like G3P[8] rotavirus A in Brazil between 2015 and 2021.Microbiol Spectr. 2024 Apr 2;12(4):e0370923. doi: 10.1128/spectrum.03709-23. Epub 2024 Mar 7. Microbiol Spectr. 2024. PMID: 38451227 Free PMC article.
-
Emergence of Equine-like G3P[8] Rotavirus Strains Infecting Children in Venezuela.Viruses. 2025 Mar 13;17(3):410. doi: 10.3390/v17030410. Viruses. 2025. PMID: 40143336 Free PMC article.
References
-
- King AM, Lefkowitz E, Adams MJ, Carstens EB, editors. Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruses. Elsevier; 2011.
-
- Harrison SC, Dormitzer PR. Structure and Function of the Rotavirus Particle. In: Svensson L, Desselberger U, editors. Viral Gastroenteritis. Academic Press; 2016. pp. 89–102.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

