SAC1 regulates autophagosomal phosphatidylinositol-4-phosphate for xenophagy-directed bacterial clearance
- PMID: 34320354
- PMCID: PMC8327279
- DOI: 10.1016/j.celrep.2021.109434
SAC1 regulates autophagosomal phosphatidylinositol-4-phosphate for xenophagy-directed bacterial clearance
Abstract
Phosphoinositides are important molecules in lipid signaling, membrane identity, and trafficking that are spatiotemporally controlled by factors from both mammalian cells and intracellular pathogens. Here, using small interfering RNA (siRNA) directed against phosphoinositide kinases and phosphatases, we screen for regulators of the host innate defense response to intracellular bacterial replication. We identify SAC1, a transmembrane phosphoinositide phosphatase, as an essential regulator of xenophagy. Depletion or inactivation of SAC1 compromises fusion between Salmonella-containing autophagosomes and lysosomes, leading to increased bacterial replication. Mechanistically, the loss of SAC1 results in aberrant accumulation of phosphatidylinositol-4-phosphate [PI(4)P] on Salmonella-containing autophagosomes, thus facilitating recruitment of SteA, a PI(4)P-binding Salmonella effector protein, which impedes lysosomal fusion. Replication of Salmonella lacking SteA is suppressed by SAC-1-deficient cells, however, demonstrating bacterial adaptation to xenophagy. Our findings uncover a paradigm in which a host protein regulates the level of its substrate and impairs the function of a bacterial effector during xenophagy.
Keywords: PI(4)P; SAC1; Salmonella; SteA; autophagosome; phosphatidylinositol; phosphoinositide phosphatase; xenophagy.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests R.J.X. is co-founder of Jnana Therapeutics and Celsius Therapeutics. These organizations had no roles in this study.
Figures
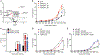
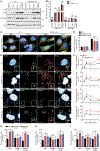
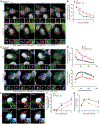

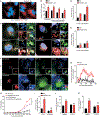

Similar articles
-
Innate host defense mechanisms SAC bacteria by regulating phosphoinositide kinases and phosphatases.Autophagy. 2022 Feb;18(2):452-454. doi: 10.1080/15548627.2021.2002102. Epub 2021 Nov 23. Autophagy. 2022. PMID: 34812102 Free PMC article.
-
PtdIns4P restriction by hydrolase SAC1 decides specific fusion of autophagosomes with lysosomes.Autophagy. 2021 Aug;17(8):1907-1917. doi: 10.1080/15548627.2020.1796321. Epub 2020 Jul 30. Autophagy. 2021. PMID: 32693712 Free PMC article.
-
Co-opted Cellular Sac1 Lipid Phosphatase and PI(4)P Phosphoinositide Are Key Host Factors during the Biogenesis of the Tombusvirus Replication Compartment.J Virol. 2020 Jun 1;94(12):e01979-19. doi: 10.1128/JVI.01979-19. Print 2020 Jun 1. J Virol. 2020. PMID: 32269127 Free PMC article.
-
A Structural View of Xenophagy, a Battle between Host and Microbes.Mol Cells. 2018 Jan 31;41(1):27-34. doi: 10.14348/molcells.2018.2274. Epub 2018 Jan 23. Mol Cells. 2018. PMID: 29370690 Free PMC article. Review.
-
Regulation of PI3K effector signalling in cancer by the phosphoinositide phosphatases.Biosci Rep. 2017 Feb 10;37(1):BSR20160432. doi: 10.1042/BSR20160432. Print 2017 Feb 28. Biosci Rep. 2017. PMID: 28082369 Free PMC article. Review.
Cited by
-
Dimethyl itaconate is effective in host-directed antimicrobial responses against mycobacterial infections through multifaceted innate immune pathways.Cell Biosci. 2023 Mar 8;13(1):49. doi: 10.1186/s13578-023-00992-x. Cell Biosci. 2023. PMID: 36882813 Free PMC article.
-
FGFR inhibition blocks NF-ĸB-dependent glucose metabolism and confers metabolic vulnerabilities in cholangiocarcinoma.Nat Commun. 2024 May 7;15(1):3805. doi: 10.1038/s41467-024-47514-y. Nat Commun. 2024. PMID: 38714664 Free PMC article.
-
Perturbation of ATG16L1 function impairs the biogenesis of Salmonella and Coxiella replication vacuoles.Mol Microbiol. 2022 Feb;117(2):235-251. doi: 10.1111/mmi.14858. Epub 2022 Jan 4. Mol Microbiol. 2022. PMID: 34874584 Free PMC article.
-
Molecular Modulators and Receptors of Selective Autophagy: Disease Implication and Identification Strategies.Int J Biol Sci. 2024 Jan 1;20(2):751-764. doi: 10.7150/ijbs.83205. eCollection 2024. Int J Biol Sci. 2024. PMID: 38169614 Free PMC article. Review.
-
A detailed molecular network map and model of the NLRP3 inflammasome.Front Immunol. 2023 Nov 15;14:1233680. doi: 10.3389/fimmu.2023.1233680. eCollection 2023. Front Immunol. 2023. PMID: 38077364 Free PMC article. Review.
References
-
- Asrat S, de Jesús DA, Hempstead AD, Ramabhadran V, and Isberg RR (2014). Bacterial pathogen manipulation of host membrane trafficking. Annu. Rev. Cell Dev. Biol 30, 79–109. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

