Development of safe and highly protective live-attenuated SARS-CoV-2 vaccine candidates by genome recoding
- PMID: 34320400
- PMCID: PMC8289629
- DOI: 10.1016/j.celrep.2021.109493
Development of safe and highly protective live-attenuated SARS-CoV-2 vaccine candidates by genome recoding
Abstract
Safe and effective vaccines are urgently needed to stop the pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). We construct a series of live attenuated vaccine candidates by large-scale recoding of the SARS-CoV-2 genome and assess their safety and efficacy in Syrian hamsters. Animals were vaccinated with a single dose of the respective recoded virus and challenged 21 days later. Two of the tested viruses do not cause clinical symptoms but are highly immunogenic and induce strong protective immunity. Attenuated viruses replicate efficiently in the upper but not in the lower airways, causing only mild pulmonary histopathology. After challenge, hamsters develop no signs of disease and rapidly clear challenge virus: at no time could infectious virus be recovered from the lungs of infected animals. The ease with which attenuated virus candidates can be produced and administered favors their further development as vaccines to combat the ongoing pandemic.
Keywords: COVID-19; Roborovski dwarf hamster; SARS-CoV-2; Syrian hamster; codon pair deoptimization; coronavirus; genome recoding; live attenuated vaccine; synthetic attenuated virus engineering.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests Freie Universität Berlin and the University of Bern received funding from a commercial partner for research similar to that described in this manuscript.
Figures

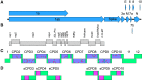
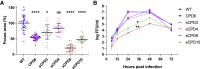


References
-
- Bazin H. A brief history of the prevention of infectious diseases by immunisations. Comp. Immunol. Microbiol. Infect. Dis. 2003;26:293–308. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

