Rapid and stable mobilization of CD8+ T cells by SARS-CoV-2 mRNA vaccine
- PMID: 34320609
- PMCID: PMC8426185
- DOI: 10.1038/s41586-021-03841-4
Rapid and stable mobilization of CD8+ T cells by SARS-CoV-2 mRNA vaccine
Abstract
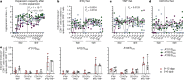
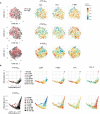
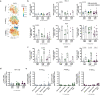
SARS-CoV-2 spike mRNA vaccines1-3 mediate protection from severe disease as early as ten days after prime vaccination3, when neutralizing antibodies are hardly detectable4-6. Vaccine-induced CD8+ T cells may therefore be the main mediators of protection at this early stage7,8. The details of their induction, comparison to natural infection, and association with other arms of vaccine-induced immunity remain, however, incompletely understood. Here we show on a single-epitope level that a stable and fully functional CD8+ T cell response is vigorously mobilized one week after prime vaccination with bnt162b2, when circulating CD4+ T cells and neutralizing antibodies are still weakly detectable. Boost vaccination induced a robust expansion that generated highly differentiated effector CD8+ T cells; however, neither the functional capacity nor the memory precursor T cell pool was affected. Compared with natural infection, vaccine-induced early memory T cells exhibited similar functional capacities but a different subset distribution. Our results indicate that CD8+ T cells are important effector cells, are expanded in the early protection window after prime vaccination, precede maturation of other effector arms of vaccine-induced immunity and are stably maintained after boost vaccination.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures











References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

