Nuclear accumulation of CHMP7 initiates nuclear pore complex injury and subsequent TDP-43 dysfunction in sporadic and familial ALS
- PMID: 34321318
- PMCID: PMC9022198
- DOI: 10.1126/scitranslmed.abe1923
Nuclear accumulation of CHMP7 initiates nuclear pore complex injury and subsequent TDP-43 dysfunction in sporadic and familial ALS
Abstract
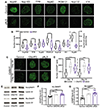
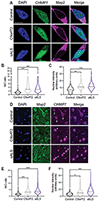
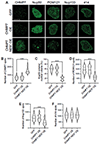
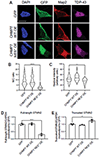
Alterations in the components [nucleoporins (Nups)] and function of the nuclear pore complex (NPC) have been implicated as contributors to the pathogenesis of genetic forms of neurodegeneration including C9orf72 amyotrophic lateral sclerosis/frontotemporal dementia (ALS/FTD). We hypothesized that Nup alterations and the consequential loss of NPC function may lie upstream of TDP-43 dysfunction and mislocalization widely observed in ALS, FTD, and related neurodegenerative diseases. Here, we provide evidence that CHMP7, a critical mediator of NPC quality control, is increased in nuclei of C9orf72 and sporadic ALS induced pluripotent stem cell (iPSC)-derived spinal neurons (iPSNs) and postmortem human motor cortex before the emergence of Nup alterations. Inhibiting the nuclear export of CHMP7 triggered Nup reduction and TDP-43 dysfunction and pathology in human neurons. Knockdown of CHMP7 alleviated disease-associated Nup alterations, deficits in Ran GTPase localization, defects in TDP-43-associated mRNA expression, and downstream glutamate-induced neuronal death. Thus, our data support a role for altered CHMP7-mediated Nup homeostasis as a prominent initiating pathological mechanism for familial and sporadic ALS and highlight the potential for CHMP7 as therapeutic target.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures

References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

