SARS-CoV-2 Vaccines Based on the Spike Glycoprotein and Implications of New Viral Variants
- PMID: 34322129
- PMCID: PMC8311925
- DOI: 10.3389/fimmu.2021.701501
SARS-CoV-2 Vaccines Based on the Spike Glycoprotein and Implications of New Viral Variants
Abstract
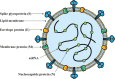
Coronavirus 19 Disease (COVID-19) originating in the province of Wuhan, China in 2019, is caused by the severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2), whose infection in humans causes mild or severe clinical manifestations that mainly affect the respiratory system. So far, the COVID-19 has caused more than 2 million deaths worldwide. SARS-CoV-2 contains the Spike (S) glycoprotein on its surface, which is the main target for current vaccine development because antibodies directed against this protein can neutralize the infection. Companies and academic institutions have developed vaccines based on the S glycoprotein, as well as its antigenic domains and epitopes, which have been proven effective in generating neutralizing antibodies. However, the emergence of new SARS-CoV-2 variants could affect the effectiveness of vaccines. Here, we review the different types of vaccines designed and developed against SARS-CoV-2, placing emphasis on whether they are based on the complete S glycoprotein, its antigenic domains such as the receptor-binding domain (RBD) or short epitopes within the S glycoprotein. We also review and discuss the possible effectiveness of these vaccines against emerging SARS-CoV-2 variants.
Keywords: RBD; SARS-CoV-2 variants; resistance to neutralization; spike glycoprotein; vaccine design.
Copyright © 2021 Martínez-Flores, Zepeda-Cervantes, Cruz-Reséndiz, Aguirre-Sampieri, Sampieri and Vaca.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- World Health Organization . WHO Director-General’s Opening Remarks at the Media Briefing on COVID-19 - 11 March 2020 Vol. 4. WHO Dir Gen speeches; (2020) Available at: https://www.who.int/dg/speeches/detail/who-director-general-s-opening-re... [Accessed May 8, 2021].
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

