The Novel Non-coding Transcriptional Regulator Gm18840 Drives Cardiomyocyte Apoptosis in Myocardial Infarction Post Ischemia/Reperfusion
- PMID: 34322480
- PMCID: PMC8312575
- DOI: 10.3389/fcell.2021.615950
The Novel Non-coding Transcriptional Regulator Gm18840 Drives Cardiomyocyte Apoptosis in Myocardial Infarction Post Ischemia/Reperfusion
Abstract
Background: Ischemia/reperfusion-mediated myocardial infarction (MIRI) is a major pathological factor implicated in the progression of ischemic heart disease, but the key factors dysregulated during MIRI have not been fully elucidated, especially those essential non-coding factors required for cardiovascular development.
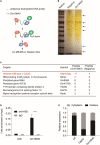
Methods: A murine MIRI model and RNA sequencing (RNA-seq) were used to identify key lncRNAs after myocardial infarction. qRT-PCR was used to validate expression in cardiac muscle tissues and myocardial cells. The role of Gm18840 in HL-1 cell growth was determined by flow cytometry experiments in vitro. Full-length Gm18840 was identified by using a rapid amplification of cDNA ends (RACE) assay. The subcellular distribution of Gm18840 was examined by nuclear/cytoplasmic RNA fractionation and qRT-PCR. RNA pulldown and RNA immunoprecipitation (RIP)-qPCR assays were performed to identify Gm18840-interacting proteins. Chromatin isolation by RNA purification (ChIRP)-seq (chromatin isolation by RNA purification) was used to identify the genome-wide binding of Gm18840 to chromatin. The regulatory activity of Gm18840 in transcriptional regulation was examined by a luciferase reporter assay and qRT-PCR.
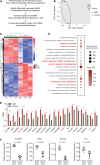
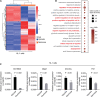
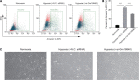
Results: Gm18840 was upregulated after myocardial infarction in both in vivo and in vitro MIRI models. Gm18840 was 1,471 nt in length and localized in both the cytoplasm and the nucleus of HL-1 cells. Functional studies showed that the knockdown of Gm18840 promoted the apoptosis of HL-1 cells. Gm18840 directly interacts with histones, including H2B, highlighting a potential function in transcriptional regulation. Further ChIRP-seq and RNA-seq analyses showed that Gm18840 is directly bound to the cis-regulatory regions of genes involved in developmental processes, such as Junb, Rras2, and Bcl3.
Conclusion: Gm18840, a novel transcriptional regulator, promoted the apoptosis of myocardial cells via direct transcriptional regulation of essential genes and might serve as a novel therapeutic target for MIRI.
Keywords: Gm18840; cardiomyocytes apoptosis; lncRNA; myocardial infarction; transcriptional regulation.
Copyright © 2021 Luo, Xiong, Huang, Deng, Zhang, Chen, Yang and Ke.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

