To Join or Not to Join: Decision Points Along the Pathway to Double-Strand Break Repair vs. Chromosome End Protection
- PMID: 34322492
- PMCID: PMC8311741
- DOI: 10.3389/fcell.2021.708763
To Join or Not to Join: Decision Points Along the Pathway to Double-Strand Break Repair vs. Chromosome End Protection
Abstract
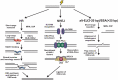
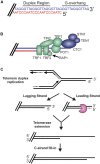
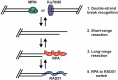
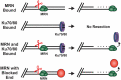
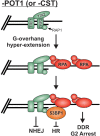
The regulation of DNA double-strand breaks (DSBs) and telomeres are diametrically opposed in the cell. DSBs are considered one of the most deleterious forms of DNA damage and must be quickly recognized and repaired. Telomeres, on the other hand, are specialized, stable DNA ends that must be protected from recognition as DSBs to inhibit unwanted chromosome fusions. Decisions to join DNA ends, or not, are therefore critical to genome stability. Yet, the processing of telomeres and DSBs share many commonalities. Accordingly, key decision points are used to shift DNA ends toward DSB repair vs. end protection. Additionally, DSBs can be repaired by two major pathways, namely homologous recombination (HR) and non-homologous end joining (NHEJ). The choice of which repair pathway is employed is also dictated by a series of decision points that shift the break toward HR or NHEJ. In this review, we will focus on these decision points and the mechanisms that dictate end protection vs. DSB repair and DSB repair choice.
Keywords: DNA repair; double-strand break; homologous recombination; non-homologous end joining; telomeres.
Copyright © 2021 Ackerson, Romney, Schuck and Stewart.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Initiation of DNA double strand break repair: signaling and single-stranded resection dictate the choice between homologous recombination, non-homologous end-joining and alternative end-joining.Am J Cancer Res. 2012;2(3):249-68. Epub 2012 Apr 21. Am J Cancer Res. 2012. PMID: 22679557 Free PMC article.
-
Shepherding DNA ends: Rif1 protects telomeres and chromosome breaks.Microb Cell. 2018 May 17;5(7):327-343. doi: 10.15698/mic2018.07.639. Microb Cell. 2018. PMID: 29992129 Free PMC article. Review.
-
Regulation of repair pathway choice at two-ended DNA double-strand breaks.Mutat Res. 2017 Oct;803-805:51-55. doi: 10.1016/j.mrfmmm.2017.07.011. Epub 2017 Jul 29. Mutat Res. 2017. PMID: 28781144 Review.
-
Mechanisms of DNA double strand break repair and chromosome aberration formation.Cytogenet Genome Res. 2004;104(1-4):14-20. doi: 10.1159/000077461. Cytogenet Genome Res. 2004. PMID: 15162010 Review.
-
The DNA damage response at dysfunctional telomeres, and at interstitial and subtelomeric DNA double-strand breaks.Genes Genet Syst. 2018 Jan 20;92(3):135-152. doi: 10.1266/ggs.17-00014. Epub 2017 Nov 22. Genes Genet Syst. 2018. PMID: 29162774 Review.
Cited by
-
Kinome-wide screening uncovers a role for Bromodomain Protein 3 in DNA double-stranded break repair.DNA Repair (Amst). 2023 Feb;122:103445. doi: 10.1016/j.dnarep.2022.103445. Epub 2022 Dec 24. DNA Repair (Amst). 2023. PMID: 36608404 Free PMC article.
-
Gastric Cancer Risk and Pathogenesis in BRCA1 and BRCA2 Carriers.Cancers (Basel). 2022 Dec 1;14(23):5953. doi: 10.3390/cancers14235953. Cancers (Basel). 2022. PMID: 36497436 Free PMC article. Review.
-
Hornerin mediates phosphorylation of the polo-box domain in Plk1 by Chk1 to induce death in mitosis.Cell Death Differ. 2023 Sep;30(9):2151-2166. doi: 10.1038/s41418-023-01208-y. Epub 2023 Aug 18. Cell Death Differ. 2023. PMID: 37596441 Free PMC article.
-
DNA-PK and ATM drive phosphorylation signatures that antagonistically regulate cytokine responses to herpesvirus infection or DNA damage.Cell Syst. 2024 Apr 17;15(4):339-361.e8. doi: 10.1016/j.cels.2024.03.003. Epub 2024 Apr 8. Cell Syst. 2024. PMID: 38593799 Free PMC article.
-
Weaponizing CRISPR/Cas9 for selective elimination of cells with an aberrant genome.DNA Repair (Amst). 2025 May;149:103840. doi: 10.1016/j.dnarep.2025.103840. Epub 2025 Apr 26. DNA Repair (Amst). 2025. PMID: 40319546 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources

