Reduced antibody cross-reactivity following infection with B.1.1.7 than with parental SARS-CoV-2 strains
- PMID: 34323691
- PMCID: PMC8352583
- DOI: 10.7554/eLife.69317
Reduced antibody cross-reactivity following infection with B.1.1.7 than with parental SARS-CoV-2 strains
Abstract
Background: The degree of heterotypic immunity induced by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) strains is a major determinant of the spread of emerging variants and the success of vaccination campaigns, but remains incompletely understood.
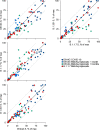
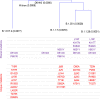
Methods: We examined the immunogenicity of SARS-CoV-2 variant B.1.1.7 (Alpha) that arose in the United Kingdom and spread globally. We determined titres of spike glycoprotein-binding antibodies and authentic virus neutralising antibodies induced by B.1.1.7 infection to infer homotypic and heterotypic immunity.
Results: Antibodies elicited by B.1.1.7 infection exhibited significantly reduced recognition and neutralisation of parental strains or of the South Africa variant B.1.351 (Beta) than of the infecting variant. The drop in cross-reactivity was significantly more pronounced following B.1.1.7 than parental strain infection.
Conclusions: The results indicate that heterotypic immunity induced by SARS-CoV-2 variants is asymmetric.
Funding: This work was supported by the Francis Crick Institute and the Max Planck Institute for Dynamics of Complex Technical Systems, Magdeburg.
Keywords: COVID-19; SARS-CoV-2; epidemiology; global health; human; infectious disease; microbiology; viral variant; virus.
© 2021, Faulkner et al.
Conflict of interest statement
NF, KN, MW, RH, MM, SP, CH, SH, MG, WB, SW, JH, HR, MS, DF, MB, Td, AS, SK, CS, SG, RB, SG, JM, RD, MH, DB, EN, GK No competing interests declared
Figures











References
-
- Aitken J, Ambrose K, Barrell S, Beale R, Bineva-Todd G, Biswas D, Byrne R, Caidan S, Cherepanov P, Churchward L, Clark G, Crawford M, Cubitt L, Dearing V, Earl C, Edwards A, Ekin C, Fidanis E, Gaiba A, Gamblin S, Gandhi S, Goldman J, Goldstone R, Grant PR, Greco M, Heaney J, Hindmarsh S, Houlihan CF, Howell M, Hubank M, Hughes D, Instrell R, Jackson D, Jamal-Hanjani M, Jiang M, Johnson M, Jones L, Kanu N, Kassiotis G, Kirk S, Kjaer S, Levett A, Levett L, Levi M, Lu WT, MacRae JI, Matthews J, McCoy LE, Moore C, Moore D, Nastouli E, Nicod J, Nightingale L, Olsen J, O'Reilly N, Pabari A, Papayannopoulos V, Patel N, Peat N, Pollitt M, Ratcliffe P, Reis E Sousa C, Rosa A, Rosenthal R, Roustan C, Rowan A, Shin GY, Snell DM, Song OR, Spyer MJ, Strange A, Swanton C, Turner JMA, Turner M, Wack A, Walker PA, Ward S, Wong WK, Wright J, Wu M, Crick COVID-19 Consortium Scalable and robust SARS-CoV-2 testing in an academic center. Nature Biotechnology. 2020;38:927–931. doi: 10.1038/s41587-020-0588-y. - DOI - PubMed
-
- Brown JC, Goldhill DH, Zhou J, Peacock TP, Frise R, Goonawardane N, Baillon L, Kugathasan R, Pinto A, McKay PF, Hassard J, Moshe M, Singanayagam A, Burgoyne T, Barclay WS. Increased transmission of Sars-Cov-2 lineage b.1.1.7 (Voc 2020212/01) Is not accounted for by a replicative advantage in primary airway cells or antibody escape. bioRxiv. 2021 doi: 10.1101/2021.02.24.432576. - DOI
-
- Cele S, Gazy I, Jackson L, Hwa SH, Tegally H, Lustig G, Giandhari J, Pillay S, Wilkinson E, Naidoo Y, Karim F, Ganga Y, Khan K, Bernstein M, Balazs AB, Gosnell BI, Hanekom W, Moosa MS, Lessells RJ, de Oliveira T, Sigal A, Network for Genomic Surveillance in South Africa. COMMIT-KZN Team Escape of SARS-CoV-2 501Y.V2 from neutralization by convalescent plasma. Nature. 2021;593:142–146. doi: 10.1038/s41586-021-03471-w. - DOI - PMC - PubMed
-
- Diamond M, Chen R, Xie X, Case J, Zhang X, VanBlargan L, Liu Y, Liu J, Errico J, Winkler E, Suryadevara N, Tahan S, Turner J, Kim W, Schmitz A, Thapa M, Wang D, Boon A, Pinto D, Presti R. Sars-Cov-2 variants show resistance to neutralization by many monoclonal and Serum-Derived polyclonal antibodies. Research Square. 2021;5:228079. doi: 10.21203/rs.3.rs-228079/v. - DOI - PMC - PubMed
-
- Edara VV, Norwood C, Floyd K, Lai L, Davis-Gardner ME, Hudson WH, Mantus G, Nyhoff LE, Adelman MW, Fineman R, Patel S, Byram R, Gomes DN, Michael G, Abdullahi H, Beydoun N, Panganiban B, McNair N, Hellmeister K, Pitts J. Reduced binding and neutralization of infection- and Vaccine-Induced antibodies to the b.1.351 (South african) Sars-Cov-2 variant. bioRxiv. 2021 doi: 10.1101/2021.02.20.432046. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

