An alternative approach for bioanalytical assay optimization for wastewater-based epidemiology of SARS-CoV-2
- PMID: 34323818
- PMCID: PMC8152210
- DOI: 10.1016/j.scitotenv.2021.148043
An alternative approach for bioanalytical assay optimization for wastewater-based epidemiology of SARS-CoV-2
Abstract
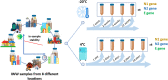
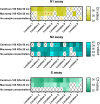
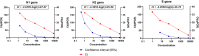
Wastewater-based epidemiology of SARS-CoV-2 could play a role in monitoring the spread of the virus in the population and controlling possible outbreaks. However, sensitive sample preparation and detection methods are necessary to detect trace levels of SARS-CoV-2 RNA in influent wastewater (IWW). Unlike predecessors, method optimization of a SARS-CoV-2 RNA concentration and detection procedure was performed with IWW samples with high viral SARS-CoV-2 RNA loads. This is of importance since the SARS-CoV-2 genome in IWW might have already been subject to in-sewer degradation into smaller genome fragments or might be present in a different form (e.g. cell debris, …). Centricon Plus-70 (100 kDa) centrifugal filter devices resulted in the lowest and most reproducible Ct-values for SARS-CoV-2 RNA. Lowering the molecular weight cut-off did not improve our limit of detection and quantification (approximately 100 copies/μL for all genes). Quantitative polymerase chain reaction (qPCR) was employed for the amplification of the N1, N2, N3 and E-gene fragments. This is one of the first studies to apply digital polymerase chain reaction (dPCR) for the detection of SARS-CoV-2 RNA in IWW. dPCR showed high variability at low concentration levels (100 copies/μL), indicating that variability in bioanalytical methods for wastewater-based epidemiology of SARS-CoV-2 might be substantial. dPCR results in IWW were in line with the results found with qPCR. On average, the N2-gene fragment showed high in-sample stability in IWW for 10 days of storage at 4 °C. Between-sample variability was substantial due to the low native concentrations in IWW. Additionally, the E-gene fragment proved to be less stable compared to the N2-gene fragment and showed higher variability. Freezing the IWW samples resulted in a 10-fold decay of loads of the N2- and E-gene fragment in IWW.
Keywords: Digital polymerase chain reaction; In-sample stability; SARS-CoV-2; Ultrafiltration; Wastewater-based epidemiology.
Copyright © 2021 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




References
-
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O'Brien J.W.…Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. - DOI - PMC - PubMed
-
- Ahmed W., Bertsch P.M., Bivins A., Bibby K., Farkas K., Gathercole A.…Kitajima M. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139960. - DOI - PMC - PubMed
-
- Ahmed W., Bivins A., Bertsch P.M., Bibby K., Choi P.M., Farkas K.…Mueller J.F. Surveillance of SARS-CoV-2 RNA in wastewater: methods optimisation and quality control are crucial for generating reliable public health information. Curr. Opin. Environ. Sci. Health. 2020 doi: 10.1016/j.coesh.2020.09.003. - DOI - PMC - PubMed
-
- Ahmed W., Tscharke B., Bertsch P.M., Bibby K., Bivins A., Choi P.…Mueller J.F. SARS-CoV-2 RNA monitoring in wastewater as a potential early warning system for COVID-19 transmission in the community: a temporal case study. Sci. Total Environ. 2021;761 doi: 10.1016/j.scitotenv.2020.144216. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

