Evolution of Renal-Disease Factor APOL1 Results in Cis and Trans Orientations at the Endoplasmic Reticulum That Both Show Cytotoxic Effects
- PMID: 34323996
- PMCID: PMC8557400
- DOI: 10.1093/molbev/msab220
Evolution of Renal-Disease Factor APOL1 Results in Cis and Trans Orientations at the Endoplasmic Reticulum That Both Show Cytotoxic Effects
Abstract
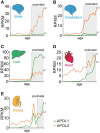
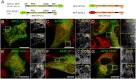
The recent and exclusively in humans and a few other higher primates expressed APOL1 (apolipoprotein L1) gene is linked to African human trypanosomiasis (also known as African sleeping sickness) as well as to different forms of kidney diseases. Whereas APOL1's role as a trypanolytic factor is well established, pathobiological mechanisms explaining its cytotoxicity in renal cells remain unclear. In this study, we compared the APOL family members using a combination of evolutionary studies and cell biological experiments to detect unique features causal for APOL1 nephrotoxic effects. We investigated available primate and mouse genome and transcriptome data to apply comparative phylogenetic and maximum likelihood selection analyses. We suggest that the APOL gene family evolved early in vertebrates and initial splitting occurred in ancestral mammals. Diversification and differentiation of functional domains continued in primates, including developing the two members APOL1 and APOL2. Their close relationship could be diagnosed by sequence similarity and a shared ancestral insertion of an AluY transposable element. Live-cell imaging analyses showed that both expressed proteins show a strong preference to localize at the endoplasmic reticulum (ER). However, glycosylation and secretion assays revealed that-unlike APOL2-APOL1 membrane insertion or association occurs in different orientations at the ER, with the disease-associated mutants facing either the luminal (cis) or cytoplasmic (trans) side of the ER. The various pools of APOL1 at the ER offer a novel perspective in explaining the broad spectrum of its observed toxic effects.
Keywords: APOL gene family, APOL phylogeny, APOL selection analyses; APOL1; APOL2; endoplasmic reticulum; evolutionary medicine; kidney disease.
© The Author(s) 2021. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures





References
-
- Almagro Armenteros JJ, Tsirigos KD, S�by CK, Petersen T N, Winther O, Brunak S, von Heijne G, Nielsen H. 2019. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol. 37(4):420–423. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

