Protein oligomer modeling guided by predicted interchain contacts in CASP14
- PMID: 34324224
- PMCID: PMC8616806
- DOI: 10.1002/prot.26197
Protein oligomer modeling guided by predicted interchain contacts in CASP14
Abstract
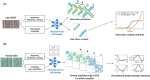
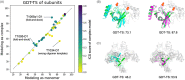
For CASP14, we developed deep learning-based methods for predicting homo-oligomeric and hetero-oligomeric contacts and used them for oligomer modeling. To build structure models, we developed an oligomer structure generation method that utilizes predicted interchain contacts to guide iterative restrained minimization from random backbone structures. We supplemented this gradient-based fold-and-dock method with template-based and ab initio docking approaches using deep learning-based subunit predictions on 29 assembly targets. These methods produced oligomer models with summed Z-scores 5.5 units higher than the next best group, with the fold-and-dock method having the best relative performance. Over the eight targets for which this method was used, the best of the five submitted models had average oligomer TM-score of 0.71 (average oligomer TM-score of the next best group: 0.64), and explicit modeling of inter-subunit interactions improved modeling of six out of 40 individual domains (ΔGDT-TS > 2.0).
Keywords: deep learning; interchain contact prediction; protein complex structure prediction; protein-protien docking.
© 2021 The Authors. Proteins: Structure, Function, and Bioinformatics published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Berggård T, Linse S, James P. Methods for the detection and analysis of protein‐protein interactions. Proteomics. 2007;7(16):2833‐2842. - PubMed
-
- Keskin O, Tuncbag N, Gursoy A. Predicting protein‐protein interactions from the molecular to the proteome level. Chem Rev. 2016;116(8):4884‐4909. - PubMed
-
- Goodsell DS, Olson AJ. Structural symmetry and protein function. Annu Rev Biophys Biomol Struct. 2000;29:105‐153. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

