Integrative analysis of genomic variants reveals new associations of candidate haploinsufficient genes with congenital heart disease
- PMID: 34324492
- PMCID: PMC8354477
- DOI: 10.1371/journal.pgen.1009679
Integrative analysis of genomic variants reveals new associations of candidate haploinsufficient genes with congenital heart disease
Erratum in
-
Correction: Integrative analysis of genomic variants reveals new associations of candidate haploinsufficient genes with congenital heart disease.PLoS Genet. 2021 Sep 21;17(9):e1009809. doi: 10.1371/journal.pgen.1009809. eCollection 2021 Sep. PLoS Genet. 2021. PMID: 34547032 Free PMC article.
Abstract
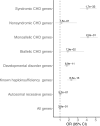
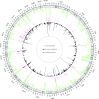
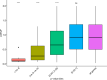
Numerous genetic studies have established a role for rare genomic variants in Congenital Heart Disease (CHD) at the copy number variation (CNV) and de novo variant (DNV) level. To identify novel haploinsufficient CHD disease genes, we performed an integrative analysis of CNVs and DNVs identified in probands with CHD including cases with sporadic thoracic aortic aneurysm. We assembled CNV data from 7,958 cases and 14,082 controls and performed a gene-wise analysis of the burden of rare genomic deletions in cases versus controls. In addition, we performed variation rate testing for DNVs identified in 2,489 parent-offspring trios. Our analysis revealed 21 genes which were significantly affected by rare CNVs and/or DNVs in probands. Fourteen of these genes have previously been associated with CHD while the remaining genes (FEZ1, MYO16, ARID1B, NALCN, WAC, KDM5B and WHSC1) have only been associated in small cases series or show new associations with CHD. In addition, a systems level analysis revealed affected protein-protein interaction networks involved in Notch signaling pathway, heart morphogenesis, DNA repair and cilia/centrosome function. Taken together, this approach highlights the importance of re-analyzing existing datasets to strengthen disease association and identify novel disease genes and pathways.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: The Department of Molecular and Human Genetics at Baylor College of Medicine receives revenue from clinical genetic testing conducted at Baylor Genetics Laboratories. M.E.H. is a co-founder of, consultant to and holds shares in Congenica, a genetics diagnostic company.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

