DogCatcher allows loop-friendly protein-protein ligation
- PMID: 34324879
- PMCID: PMC8878318
- DOI: 10.1016/j.chembiol.2021.07.005
DogCatcher allows loop-friendly protein-protein ligation
Abstract
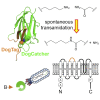
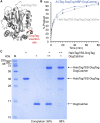
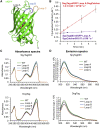
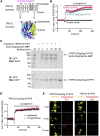
There are many efficient ways to connect proteins at termini. However, connecting at a loop is difficult because of lower flexibility and variable environment. Here, we have developed DogCatcher, a protein that forms a spontaneous isopeptide bond with DogTag peptide. DogTag/DogCatcher was generated initially by splitting a Streptococcus pneumoniae adhesin. We optimized DogTag/DogCatcher through rational design and evolution, increasing reaction rate by 250-fold and establishing millimolar solubility of DogCatcher. When fused to a protein terminus, DogTag/DogCatcher reacts slower than SpyTag003/SpyCatcher003. However, inserted in loops of a fluorescent protein or enzyme, DogTag reacts much faster than SpyTag003. Like many membrane proteins, the ion channel TRPC5 has no surface-exposed termini. DogTag in a TRPC5 extracellular loop allowed normal calcium flux and specific covalent labeling on cells in 1 min. DogTag/DogCatcher reacts under diverse conditions, at nanomolar concentrations, and to 98% conversion. Loop-friendly ligation should expand the toolbox for creating protein architectures.
Keywords: SpyTag; TRPC; bioconjugation; chemical biology; epitope tag; ion channel; protein design; protein engineering; split protein; synthetic biology.
Copyright © 2021 The Authors. Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of interests M.H. and J.H. are authors on a patent covering RrgA splitting (UK Intellectual Property Office 1509782.7). M.H. is an author on a patent covering DogTag and SnoopLigase (UK Intellectual Property Office 1705750.6). M.H. and A.H.K. are authors on a patent application for SpyTag003/SpyCatcher003 (UK Intellectual Property Office, 1903479.2). M.H., A.H.K., M.P.F., and V.K.Y. are authors on a patent application related to DogCatcher (UK Intellectual Property Office 2104999.4.). M.H. is a SpyBiotech co-founder, shareholder, and consultant. All other authors have no conflicts of interest.
Figures




References
-
- Anderson P.A., Greenberg R.M. Phylogeny of ion channels: clues to structure and function. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2001;129:17–28. - PubMed
-
- Bae Y., Lee S.K., Chae Y.C., Park C.Y., Kang S. Accessibility-dependent topology studies of membrane proteins using a SpyTag/SpyCatcher protein-ligation system. Int. J. Biol. Macromol. 2021;175:171–178. - PubMed
-
- Banerjee A., Howarth M. Nanoteamwork: covalent protein assembly beyond duets towards protein ensembles and orchestras. Curr. Opin. Biotechnol. 2018;51:16–23. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

