Temporal shifts in antibiotic resistance elements govern phage-pathogen conflicts
- PMID: 34326207
- PMCID: PMC9064180
- DOI: 10.1126/science.abg2166
Temporal shifts in antibiotic resistance elements govern phage-pathogen conflicts
Abstract
Bacteriophage predation selects for diverse antiphage systems that frequently cluster on mobilizable defense islands in bacterial genomes. However, molecular insight into the reciprocal dynamics of phage-bacterial adaptations in nature is lacking, particularly in clinical contexts where there is need to inform phage therapy efforts and to understand how phages drive pathogen evolution. Using time-shift experiments, we uncovered fluctuations in Vibrio cholerae's resistance to phages in clinical samples. We mapped phage resistance determinants to SXT integrative and conjugative elements (ICEs), which notoriously also confer antibiotic resistance. We found that SXT ICEs, which are widespread in γ-proteobacteria, invariably encode phage defense systems localized to a single hotspot of genetic exchange. We identified mechanisms that allow phage to counter SXT-mediated defense in clinical samples, and document the selection of a novel phage-encoded defense inhibitor. Phage infection stimulates high-frequency SXT ICE conjugation, leading to the concurrent dissemination of phage and antibiotic resistances.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Supplementary Materials:
Figs. S1 to S14
Tables S1 to S4
Other Supplementary Materials for this manuscript include the following:
Data S1 [Strains used in this study]
Data S2 [SXT ICE hotspot 5 contents]
Data S3 [
Data S4 [ICP1 isolates sequenced in this study]
Data S5 [Accession numbers for SXT ICEs used for BLAST analysis]
Data S6 [SXT ICEs and hotspot 5 returned by queried sequences from BLAST analysis]
Data S7 [Recognition motifs in phages and MGEs tested]
Figures


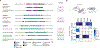

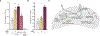
References
-
- Wilhelm SW, a Suttle C, Viruses and Nutrient Cycles in the Sea aquatic food webs. Bioscience. 49, 781–788 (1999).
-
- Koskella B, New approaches to characterizing bacteria–phage interactions in microbial communities and microbiomes. Environ. Microbiol. Rep. 11, 15–16 (2019). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

