Replication-dependent histone biosynthesis is coupled to cell-cycle commitment
- PMID: 34326254
- PMCID: PMC8346857
- DOI: 10.1073/pnas.2100178118
Replication-dependent histone biosynthesis is coupled to cell-cycle commitment
Abstract
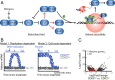
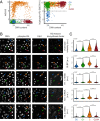
The current model of replication-dependent (RD) histone biosynthesis posits that RD histone gene expression is coupled to DNA replication, occurring only in S phase of the cell cycle once DNA synthesis has begun. However, several key factors in the RD histone biosynthesis pathway are up-regulated by E2F or phosphorylated by CDK2, suggesting these processes may instead begin much earlier, at the point of cell-cycle commitment. In this study, we use both fixed- and live-cell imaging of human cells to address this question, revealing a hybrid model in which RD histone biosynthesis is first initiated in G1, followed by a strong increase in histone production in S phase of the cell cycle. This suggests a mechanism by which cells that have committed to the cell cycle build up an initial small pool of RD histones to be available for the start of DNA replication, before producing most of the necessary histones required in S phase. Thus, a clear distinction exists at completion of mitosis between cells that are born with the intention of proceeding through the cell cycle and replicating their DNA and cells that have chosen to exit the cell cycle and have no immediate need for histone synthesis.
Keywords: NPAT; SLBP; histone locus body; replication-dependent histone; restriction point.
Copyright © 2021 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

