Differential immune responses in pregnant patients recovered from COVID-19
- PMID: 34326311
- PMCID: PMC8320317
- DOI: 10.1038/s41392-021-00703-3
Differential immune responses in pregnant patients recovered from COVID-19
Abstract
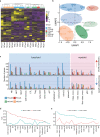
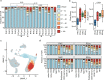
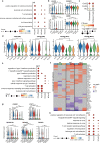
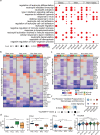
Pregnant women are generally more susceptible to viral infection. Although the impact of SARS-CoV-2 in pregnancy remains to be determined, evidence indicates that the risk factors for severe COVID-19 are similar in pregnancy to the general population. Here we systemically analyzed the clinical characteristics of pregnant and non-pregnant female COVID-19 patients who were hospitalized during the same period and found that pregnant patients developed marked lymphopenia and higher inflammation evident by higher C-reactive protein and IL-6. To elucidate the pathways that might contribute to immunopathology or protective immunity against COVID-19 during pregnancy, we applied single-cell mRNA sequencing to profile peripheral blood mononuclear cells from four pregnant and six non-pregnant female patients after recovery along with four pregnant and three non-pregnant healthy donors. We found normal clonal expansion of T cells in the pregnant patients, heightened activation and chemotaxis in NK, NKT, and MAIT cells, and differential interferon responses in the monocyte compartment. Our data present a unique feature in both innate and adaptive immune responses in pregnant patients recovered from COVID-19.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Immune Response to COVID-19 During Pregnancy.Front Immunol. 2021 May 3;12:675476. doi: 10.3389/fimmu.2021.675476. eCollection 2021. Front Immunol. 2021. PMID: 34012458 Free PMC article.
-
SARS-CoV-2 infection elucidates features of pregnancy-specific immunity.Cell Rep. 2024 Nov 26;43(11):114933. doi: 10.1016/j.celrep.2024.114933. Epub 2024 Nov 5. Cell Rep. 2024. PMID: 39504241 Free PMC article.
-
T Lymphocyte Characteristic Changes Under Serum Cytokine Deviations and Prognostic Factors of COVID-19 in Pregnant Women.Appl Biochem Biotechnol. 2024 Jul;196(7):4366-4381. doi: 10.1007/s12010-023-04775-5. Epub 2023 Nov 10. Appl Biochem Biotechnol. 2024. PMID: 37947946
-
Comprehensive Analysis of the ILCs and Unconventional T Cells in Virus Infection: Profiling and Dynamics Associated with COVID-19 Disease for a Future Monitoring System and Therapeutic Opportunities.Cells. 2022 Feb 4;11(3):542. doi: 10.3390/cells11030542. Cells. 2022. PMID: 35159352 Free PMC article. Review.
-
What immunological and hormonal protective factors lower the risk of COVID-19 related deaths in pregnant women?J Reprod Immunol. 2020 Nov;142:103180. doi: 10.1016/j.jri.2020.103180. Epub 2020 Jul 18. J Reprod Immunol. 2020. PMID: 32739645 Free PMC article. Review.
Cited by
-
The effect of SARS-CoV-2 infection and vaccination on Th17 and regulatory T cells in a pregnancy cohort in NYC.Front Immunol. 2024 Mar 5;15:1350288. doi: 10.3389/fimmu.2024.1350288. eCollection 2024. Front Immunol. 2024. PMID: 38504979 Free PMC article.
-
Single-cell RNA-sequencing highlights a curtailed NK cell function in convalescent COVID-19 pregnant women.Front Immunol. 2025 Jun 30;16:1560391. doi: 10.3389/fimmu.2025.1560391. eCollection 2025. Front Immunol. 2025. PMID: 40661959 Free PMC article.
-
Dynamics of Innate Immunity in SARS-CoV-2 Infections: Exploring the Impact of Natural Killer Cells, Inflammatory Responses, Viral Evasion Strategies, and Severity.Cells. 2025 May 22;14(11):763. doi: 10.3390/cells14110763. Cells. 2025. PMID: 40497938 Free PMC article. Review.
-
Mucosal-associated invariant T cells in infectious diseases of respiratory system: recent advancements and applications.J Inflamm (Lond). 2024 Feb 28;21(1):6. doi: 10.1186/s12950-024-00376-z. J Inflamm (Lond). 2024. PMID: 38419084 Free PMC article. Review.
-
Immune and cytokine alterations and RNA-sequencing analysis in gestational tissues from pregnant women after recovery from COVID-19.BMC Infect Dis. 2023 Sep 21;23(1):620. doi: 10.1186/s12879-023-08607-z. BMC Infect Dis. 2023. PMID: 37735363 Free PMC article.
References
-
- WHO. Coronavirus disease (COVID-19) pandemic. https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (2021).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

