SLITRK5 is a negative regulator of hedgehog signaling in osteoblasts
- PMID: 34326333
- PMCID: PMC8322311
- DOI: 10.1038/s41467-021-24819-w
SLITRK5 is a negative regulator of hedgehog signaling in osteoblasts
Abstract
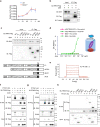
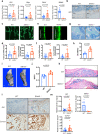
Hedgehog signaling is essential for bone formation, including functioning as a means for the growth plate to drive skeletal mineralization. However, the mechanisms regulating hedgehog signaling specifically in bone-forming osteoblasts are largely unknown. Here, we identified SLIT and NTRK-like protein-5(Slitrk5), a transmembrane protein with few identified functions, as a negative regulator of hedgehog signaling in osteoblasts. Slitrk5 is selectively expressed in osteoblasts and loss of Slitrk5 enhanced osteoblast differentiation in vitro and in vivo. Loss of SLITRK5 in vitro leads to increased hedgehog signaling and overexpression of SLITRK5 in osteoblasts inhibits the induction of targets downstream of hedgehog signaling. Mechanistically, SLITRK5 binds to hedgehog ligands via its extracellular domain and interacts with PTCH1 via its intracellular domain. SLITRK5 is present in the primary cilium, and loss of SLITRK5 enhances SMO ciliary enrichment upon SHH stimulation. Thus, SLITRK5 is a negative regulator of hedgehog signaling in osteoblasts that may be attractive as a therapeutic target to enhance bone formation.
© 2021. The Author(s).
Conflict of interest statement
J.H.S. is a scientific co-founder of AAVAA therapeutics and holds equity in this company. The remaining authors declare no competing interests.
Figures



References
-
- Ganesan K., Jandu J. S., Roane D. Osteoporosis (Secondary). In StatPearls (2020). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

