Single-cell transcriptomics reveals heterogeneous progression and EGFR activation in pancreatic adenosquamous carcinoma
- PMID: 34326696
- PMCID: PMC8315026
- DOI: 10.7150/ijbs.58886
Single-cell transcriptomics reveals heterogeneous progression and EGFR activation in pancreatic adenosquamous carcinoma
Abstract
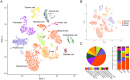
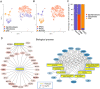
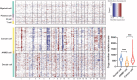
Pancreatic adenosquamous carcinoma (PASC) - a rare pathological pancreatic cancer (PC) type - has a poor prognosis due to high malignancy. To examine the heterogeneity of PASC, we performed single-cell RNA sequencing (scRNA-seq) profiling with sample tissues from a healthy donor pancreas, an intraductal papillary mucinous neoplasm, and a patient with PASC. Of 9,887 individual cells, ten cell subpopulations were identified, including myeloid, immune, ductal, fibroblast, acinar, stellate, endothelial, and cancer cells. Cancer cells were divided into five clusters. Notably, cluster 1 exhibited stem-like phenotypes expressing UBE2C, ASPM, and TOP2A. We found that S100A2 is a potential biomarker for cancer cells. LGALS1, NPM1, RACK1, and PERP were upregulated from ductal to cancer cells. Furthermore, the copy number variations in ductal and cancer cells were greater than in the reference cells. The expression of EREG, FCGR2A, CCL4L2, and CTSC increased in myeloid cells from the normal pancreas to PASC. The gene sets expressed by cancer-associated fibroblasts were enriched in the immunosuppressive pathways. We demonstrate that EGFR-associated ligand-receptor pairs are activated in ductal-stromal cell communications. Hence, this study revealed the heterogeneous variations of ductal and stromal cells, defined cancer-associated signaling pathways, and deciphered intercellular interactions following PASC progression.
Keywords: cell-cell communication; heterogeneity; intraductal papillary mucinous neoplasm; pancreatic adenosquamous carcinoma; pancreatic cancer; single-cell RNA sequencing.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures








References
-
- Kleeff J, Korc M, Apte M. et al. Pancreatic cancer. Nat Rev Dis Primers. 2016;2:16022. - PubMed
-
- Hester CA, Augustine MM, Choti MA. et al. Comparative outcomes of adenosquamous carcinoma of the pancreas: An analysis of the National Cancer Database. J Surg Oncol. 2018;118(1):21–30. - PubMed
-
- Boecker J, Feyerabend B, Tiemann K. et al. Adenosquamous Carcinoma of the Pancreas Comprise a Heterogeneous Group of Tumors With the Worst Outcome: A Clinicopathological Analysis of 25 Cases Identified in 562 Pancreatic Carcinomas Resected With Curative Intent. Pancreas. 2020;49(5):683–91. - PubMed
-
- Fang Y, Su Z, Xie J. et al. Genomic signatures of pancreatic adenosquamous carcinoma (PASC) J Pathol. 2017;243(2):155–9. - PubMed
-
- Brody JR, Costantino CL, Potoczek M. et al. Adenosquamous carcinoma of the pancreas harbors KRAS2, DPC4 and TP53 molecular alterations similar to pancreatic ductal adenocarcinoma. Mod Pathol. 2009;22(5):651–9. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

