Discovery of Small-Molecule Inhibitors of SARS-CoV-2 Proteins Using a Computational and Experimental Pipeline
- PMID: 34327214
- PMCID: PMC8315004
- DOI: 10.3389/fmolb.2021.678701
Discovery of Small-Molecule Inhibitors of SARS-CoV-2 Proteins Using a Computational and Experimental Pipeline
Abstract
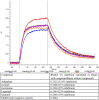
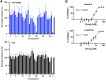
A rapid response is necessary to contain emergent biological outbreaks before they can become pandemics. The novel coronavirus (SARS-CoV-2) that causes COVID-19 was first reported in December of 2019 in Wuhan, China and reached most corners of the globe in less than two months. In just over a year since the initial infections, COVID-19 infected almost 100 million people worldwide. Although similar to SARS-CoV and MERS-CoV, SARS-CoV-2 has resisted treatments that are effective against other coronaviruses. Crystal structures of two SARS-CoV-2 proteins, spike protein and main protease, have been reported and can serve as targets for studies in neutralizing this threat. We have employed molecular docking, molecular dynamics simulations, and machine learning to identify from a library of 26 million molecules possible candidate compounds that may attenuate or neutralize the effects of this virus. The viability of selected candidate compounds against SARS-CoV-2 was determined experimentally by biolayer interferometry and FRET-based activity protein assays along with virus-based assays. In the pseudovirus assay, imatinib and lapatinib had IC50 values below 10 μM, while candesartan cilexetil had an IC50 value of approximately 67 µM against Mpro in a FRET-based activity assay. Comparatively, candesartan cilexetil had the highest selectivity index of all compounds tested as its half-maximal cytotoxicity concentration 50 (CC50) value was the only one greater than the limit of the assay (>100 μM).
Keywords: COVID-19; FRET; live virus; machine-learning; main protease; molecular simulations; protein assays; spike protein.
Copyright © 2021 Lau, Negrete, Bennett, Bennion, Borucki, Bourguet, Epstein, Franco, Harmon, He, Jones, Kim, Kirshner, Lao, Lo, McLoughlin, Mosesso, Murugesh, Saada, Segelke, Stefan, Stevenson, Torres, Weilhammer, Wong, Yang, Zemla, Zhang, Zhu, Allen and Lightstone.
Conflict of interest statement
Authors OAN, BH, RM, EAS, and MAS were employed by Sandia National Laboratories. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

