Hyperspectral reflectance-based phenotyping for quantitative genetics in crops: Progress and challenges
- PMID: 34327323
- PMCID: PMC8299078
- DOI: 10.1016/j.xplc.2021.100209
Hyperspectral reflectance-based phenotyping for quantitative genetics in crops: Progress and challenges
Abstract
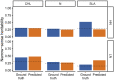
Many biochemical and physiological properties of plants that are of interest to breeders and geneticists have extremely low throughput and/or can only be measured destructively. This has limited the use of information on natural variation in nutrient and metabolite abundance, as well as photosynthetic capacity in quantitative genetic contexts where it is necessary to collect data from hundreds or thousands of plants. A number of recent studies have demonstrated the potential to estimate many of these traits from hyperspectral reflectance data, primarily in ecophysiological contexts. Here, we summarize recent advances in the use of hyperspectral reflectance data for plant phenotyping, and discuss both the potential benefits and remaining challenges to its application in plant genetics contexts. The performances of previously published models in estimating six traits from hyperspectral reflectance data in maize were evaluated on new sample datasets, and the resulting predicted trait values shown to be heritable (e.g., explained by genetic factors) were estimated. The adoption of hyperspectral reflectance-based phenotyping beyond its current uses may accelerate the study of genes controlling natural variation in biochemical and physiological traits.
Keywords: hyperspectral reflectance; maize; phenotyping; quantitative genetics.
© 2021 The Author(s).
Figures


References
-
- Bruning B., Berger B., Lewis M., Liu H., Garnett T. Approaches, applications, and future directions for hyperspectral vegetation studies: an emphasis on yield-limiting factors in wheat. Plant Phenome J. 2020;3:e20007.
-
- Buckler E.S., Holland J.B., Bradbury P.J., Acharya C.B., Brown P.J., Browne C., Ersoz E., Flint-Garcia S., Garcia A., Glaubitz J.C. The genetic architecture of maize flowering time. Science. 2009;325:714–718. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

