Discovery and Characterization of Spike N-Terminal Domain-Binding Aptamers for Rapid SARS-CoV-2 Detection
- PMID: 34328683
- PMCID: PMC8426805
- DOI: 10.1002/anie.202107730
Discovery and Characterization of Spike N-Terminal Domain-Binding Aptamers for Rapid SARS-CoV-2 Detection
Abstract
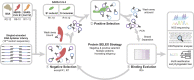
The coronavirus disease 2019 (COVID-19) pandemic has devastated families and disrupted healthcare, economies and societies across the globe. Molecular recognition agents that are specific for distinct viral proteins are critical components for rapid diagnostics and targeted therapeutics. In this work, we demonstrate the selection of novel DNA aptamers that bind to the SARS-CoV-2 spike glycoprotein with high specificity and affinity (<80 nM). Through binding assays and high resolution cryo-EM, we demonstrate that SNAP1 (SARS-CoV-2 spike protein N-terminal domain-binding aptamer 1) binds to the S N-terminal domain. We applied SNAP1 in lateral flow assays (LFAs) and ELISAs to detect UV-inactivated SARS-CoV-2 at concentrations as low as 5×105 copies mL-1 . SNAP1 is therefore a promising molecular tool for SARS-CoV-2 diagnostics.
Keywords: SARS-CoV-2; aptamers; coronavirus; cryo-EM; immunoassays.
© 2021 Wiley-VCH GmbH.
Conflict of interest statement
We have provisional patent filed on the aptamer sequences in this manuscript.
Figures


References
-
- None
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

