Molecular features and vulnerabilities of recurrent chordomas
- PMID: 34330313
- PMCID: PMC8325178
- DOI: 10.1186/s13046-021-02037-y
Molecular features and vulnerabilities of recurrent chordomas
Abstract
Background: Tumor recurrence is one of the major challenges in clinical management of chordoma. Despite R0-resection, approximately 50% of chordomas recur within ten years after initial surgery. The underlying molecular processes are poorly understood resulting in the lack of associated therapeutic options. This is not least due to the absence of appropriate cell culture models of this orphan disease.
Methods: The intra-personal progression model cell lines U-CH11 and U-CH11R were compared using array comparative genomic hybridization, expression arrays, RNA-seq, and immunocytochemistry. Cell line origin was confirmed by short tandem repeat analysis. Inter-personal cell culture models (n = 6) were examined to validate whether the new model is representative. Cell viability after HOX/PBX complex inhibition with small peptides was determined by MTS assays.
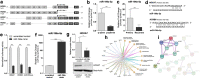
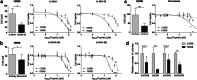
Results: Using whole genome microarray analyses, striking differences in gene expression between primary and recurrent chordomas were identified. These expression differences were confirmed in the world's first intra-personal model of chordoma relapse consisting of cell lines established from a primary (U-CH11) and the corresponding recurrent tumor (U-CH11R). Array comparative genomic hybridization and RNA-sequencing analyses revealed profound genetic similarities between both cell lines pointing to transcriptomic reprogramming as a key mechanism of chordoma progression. Network analysis of the recurrence specific genes highlighted HOX/PBX signaling as a common dysregulated event. Hence, HOX/PBX complexes were used as so far unknown therapeutic targets in recurrent chordomas. Treating chordoma cell lines with the complex formation inhibiting peptide HXR9 induced cFOS mediated apoptosis in all chordoma cell lines tested. This effect was significantly stronger in cell lines established from chordoma relapses.
Conclusion: Clearly differing gene expression patterns and vulnerabilities to HOX/PBX complex inhibition in highly therapy resistant chordoma relapses were identified using the first intra-personal loco-regional and further inter-personal chordoma progression models. For the first time, HOX/PBX interference was used to induce cell death in chordoma and might serve as the basic concept of an upcoming targeted therapy for chordomas of all progression stages.
Keywords: Apoptosis; Cell lines; Chordoma; HOX; PBX; Progression model.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- Miettinen M, Wang Z, Lasota J, Heery C, Schlom J, Palena C. Nuclear Brachyury expression is consistent in Chordoma, common in germ cell tumors and small cell carcinomas, and rare in other carcinomas and sarcomas: an Immunohistochemical study of 5229 cases. Am J Surg Pathol. 2015;39(10):1305–1312. doi: 10.1097/PAS.0000000000000462. - DOI - PMC - PubMed
-
- Sharifnia T, Wawer MJ, Chen T, Huang QY, Weir BA, Sizemore A, Lawlor MA, Goodale A, Cowley GS, Vazquez F, Ott CJ, Francis JM, Sassi S, Cogswell P, Sheppard HE, Zhang T, Gray NS, Clarke PA, Blagg J, Workman P, Sommer J, Hornicek F, Root DE, Hahn WC, Bradner JE, Wong KK, Clemons PA, Lin CY, Kotz JD, Schreiber SL. Small-molecule targeting of brachyury transcription factor addiction in chordoma. Nat Med. 2019;25(2):292–300. doi: 10.1038/s41591-018-0312-3. - DOI - PMC - PubMed
-
- Hsu W, Mohyeldin A, Shah SR, Rhys CM a, Johnson LF, Sedora-Roman NI, Kosztowski TA, Awad OA, McCarthy EF, Loeb DM, et al. Generation of chordoma cell line JHC7 and the identification of Brachyury as a novel molecular target. J Neurosurg. 2011;115(4):760–769. doi: 10.3171/2011.5.JNS11185. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

