Cytochrome P450-Catalyzed Metabolism of Cannabidiol to the Active Metabolite 7-Hydroxy-Cannabidiol
- PMID: 34330718
- PMCID: PMC11025033
- DOI: 10.1124/dmd.120.000350
Cytochrome P450-Catalyzed Metabolism of Cannabidiol to the Active Metabolite 7-Hydroxy-Cannabidiol
Abstract
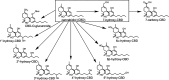
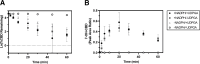
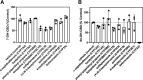
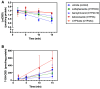
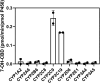
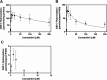
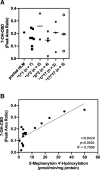
Cannabidiol (CBD) is a naturally occurring nonpsychotoxic phytocannabinoid that has gained increasing attention as a popular consumer product and for its use in Food and Drug Administration-approved Epidiolex (CBD oral solution) for the treatment of Lennox-Gastaut syndrome and Dravet syndrome. CBD was previously reported to be metabolized primarily by CYP2C19 and CYP3A4, with minor contributions from UDP-glucuronosyltransferases. 7-Hydroxy-CBD (7-OH-CBD) is the primary active metabolite with equipotent activity compared with CBD. Given the polymorphic nature of CYP2C19, we hypothesized that variable CYP2C19 expression may lead to interindividual differences in CBD metabolism to 7-OH-CBD. The objectives of this study were to further characterize the roles of cytochrome P450 enzymes in CBD metabolism, specifically to the active metabolite 7-OH-CBD, and to investigate the impact of CYP2C19 polymorphism on CBD metabolism in genotyped human liver microsomes. The results from reaction phenotyping experiments with recombinant cytochrome P450 enzymes and cytochrome P450-selective chemical inhibitors indicated that both CYP2C19 and CYP2C9 are capable of CBD metabolism to 7-OH-CBD. CYP3A played a major role in CBD metabolic clearance via oxidation at sites other than the 7-position. In genotyped human liver microsomes, 7-OH-CBD formation was positively correlated with CYP2C19 activity but was not associated with CYP2C19 genotype. In a subset of single-donor human liver microsomes with moderate to low CYP2C19 activity, CYP2C9 inhibition significantly reduced 7-OH-CBD formation, suggesting that CYP2C9 may play a greater role in CBD 7-hydroxylation than previously thought. Collectively, these data indicate that both CYP2C19 and CYP2C9 are important contributors in CBD metabolism to the active metabolite 7-OH-CBD. SIGNIFICANCE STATEMENT: This study demonstrates that both CYP2C19 and CYP2C9 are involved in CBD metabolism to the active metabolite 7-OH-CBD and that CYP3A4 is a major contributor to CBD metabolism through pathways other than 7-hydroxylation. 7-OH-CBD formation was associated with human liver microsomal CYP2C19 activity, but not CYP2C19 genotype, and CYP2C9 was found to contribute significantly to 7-OH-CBD generation. These findings have implications for patients taking CBD who may be at risk for clinically important cytochrome P450-mediated drug interactions.
Copyright © 2021 by The American Society for Pharmacology and Experimental Therapeutics.
Figures

References
-
- Achour BBarber J, Rostami-Hodjegan A (2014) Expression of hepatic drug-metabolizing cytochrome p450 enzymes and their intercorrelations: a meta-analysis. Drug Metab Dispos 42:1349–1356. - PubMed
-
- Copeland RA (2000) Enzymes: A Practical Introduction to Structure, Mechanism, and Data Analysis, Ed. 2nd. Wiley-VCH, Inc., New York.
-
- Damkier P, Lassen D, Christensen MMH, Madsen KG, Hellfritzsch M, Pottegård A(2019) Interaction between warfarin and cannabis. Basic Clin Pharmacol Toxicol 124:28–31. - PubMed
-
- Gaston TE, Bebin EM, Cutter GR, Liu Y, Szaflarski JP; UAB CBD Program (2017) Interactions between cannabidiol and commonly used antiepileptic drugs. Epilepsia 58:1586–1592. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

