Extensive variation in the intelectin gene family in laboratory and wild mouse strains
- PMID: 34330944
- PMCID: PMC8324875
- DOI: 10.1038/s41598-021-94679-3
Extensive variation in the intelectin gene family in laboratory and wild mouse strains
Abstract
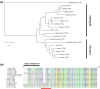
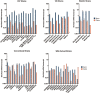
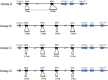
Intelectins are a family of multimeric secreted proteins that bind microbe-specific glycans. Both genetic and functional studies have suggested that intelectins have an important role in innate immunity and are involved in the etiology of various human diseases, including inflammatory bowel disease. Experiments investigating the role of intelectins in human disease using mouse models are limited by the fact that there is not a clear one-to-one relationship between intelectin genes in humans and mice, and that the number of intelectin genes varies between different mouse strains. In this study we show by gene sequence and gene expression analysis that human intelectin-1 (ITLN1) has multiple orthologues in mice, including a functional homologue Itln1; however, human intelectin-2 has no such orthologue or homologue. We confirm that all sub-strains of the C57 mouse strain have a large deletion resulting in retention of only one intelectin gene, Itln1. The majority of laboratory strains have a full complement of six intelectin genes, except CAST, SPRET, SKIVE, MOLF and PANCEVO strains, which are derived from different mouse species/subspecies and encode different complements of intelectin genes. In wild mice, intelectin deletions are polymorphic in Mus musculus castaneus and Mus musculus domesticus. Further sequence analysis shows that Itln3 and Itln5 are polymorphic pseudogenes due to premature truncating mutations, and that mouse Itln1 has undergone recent adaptive evolution. Taken together, our study shows extensive diversity in intelectin genes in both laboratory and wild-mice, suggesting a pattern of birth-and-death evolution. In addition, our data provide a foundation for further experimental investigation of the role of intelectins in disease.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
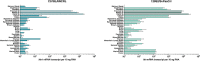


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

