The omnigenic model and polygenic prediction of complex traits
- PMID: 34331855
- PMCID: PMC8456163
- DOI: 10.1016/j.ajhg.2021.07.003
The omnigenic model and polygenic prediction of complex traits
Abstract
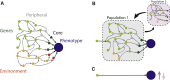
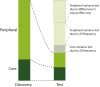
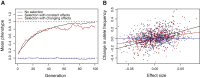
The omnigenic model was proposed as a framework to understand the highly polygenic architecture of complex traits revealed by genome-wide association studies (GWASs). I argue that this model also explains recent observations about cross-population genetic effects, specifically the low transferability of polygenic scores and the lack of clear evidence for polygenic selection. In particular, the omnigenic model explains why the effects of most GWAS variants vary between populations. This interpretation has several consequences for the evolutionary interpretation and practical use of GWAS summary statistics and polygenic scores. First, some polygenic scores may be applicable only in populations of the same ancestry and environment as the discovery population. Second, most GWAS associations will have differing effects between populations and are unlikely to be robust clinical targets. Finally, it may not always be possible to detect polygenic selection from population genetic data. These considerations make it difficult to interpret the clinical and evolutionary meanings of polygenic scores without an explicit model of genetic architecture.
Copyright © 2021 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The author declares no competing interests.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

