A Toolbox for Efficient Proximity-Dependent Biotinylation in Zebrafish Embryos
- PMID: 34332124
- PMCID: PMC8383115
- DOI: 10.1016/j.mcpro.2021.100128
A Toolbox for Efficient Proximity-Dependent Biotinylation in Zebrafish Embryos
Abstract
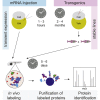
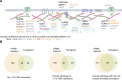
Understanding how proteins are organized in compartments is essential to elucidating their function. While proximity-dependent approaches such as BioID have enabled a massive increase in information about organelles, protein complexes, and other structures in cell culture, to date there have been only a few studies on living vertebrates. Here, we adapted proximity labeling for protein discovery in vivo in the vertebrate model organism, zebrafish. Using lamin A (LMNA) as bait and green fluorescent protein (GFP) as a negative control, we developed, optimized, and benchmarked in vivo TurboID and miniTurbo labeling in early zebrafish embryos. We developed both an mRNA injection protocol and a transgenic system in which transgene expression is controlled by a heat shock promoter. In both cases, biotin is provided directly in the egg water, and we demonstrate that 12 h of labeling are sufficient for biotinylation of prey proteins, which should permit time-resolved analysis of development. After statistical scoring, we found that the proximal partners of LMNA detected in each system were enriched for nuclear envelope and nuclear membrane proteins and included many orthologs of human proteins identified as proximity partners of lamin A in mammalian cell culture. The tools and protocols developed here will allow zebrafish researchers to complement genetic tools with powerful proteomics approaches.
Keywords: BioID; Proximity-dependent biotinylation; TurboID; miniTurbo; zebrafish embryo.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflicts of interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
-
- Lieschke G.J., Currie P.D. Animal models of human disease: Zebrafish swim into view. Nat. Rev. Genet. 2007;8:353–367. - PubMed
-
- Koster R., Sassen W.A. A molecular toolbox for genetic manipulation of zebrafish. Adv. Genomics Genet. 2015;5:151.
-
- Xin Y., Duan C. Methods in Molecular Biology. Humana Press Inc; Clifton, NJ: 2018. pp. 205–211.
-
- Felker A., Mosimann C. Contemporary zebrafish transgenesis with Tol2 and application for Cre/lox recombination experiments. Methods Cell Biol. 2016;135:219–244. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

