An ancient whole-genome duplication event and its contribution to flavor compounds in the tea plant (Camellia sinensis)
- PMID: 34333548
- PMCID: PMC8325681
- DOI: 10.1038/s41438-021-00613-z
An ancient whole-genome duplication event and its contribution to flavor compounds in the tea plant (Camellia sinensis)
Abstract
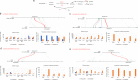
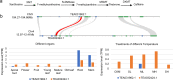
Tea, coffee, and cocoa are the three most popular nonalcoholic beverages in the world and have extremely high economic and cultural value. The genomes of four tea plant varieties have recently been sequenced, but there is some debate regarding the characterization of a whole-genome duplication (WGD) event in tea plants. Whether the WGD in the tea plant is shared with other plants in order Ericales and how it contributed to tea plant evolution remained unanswered. Here we re-analyzed the tea plant genome and provided evidence that tea experienced only WGD event after the core-eudicot whole-genome triplication (WGT) event. This WGD was shared by the Polemonioids-Primuloids-Core Ericales (PPC) sections, encompassing at least 17 families in the order Ericales. In addition, our study identified eight pairs of duplicated genes in the catechins biosynthesis pathway, four pairs of duplicated genes in the theanine biosynthesis pathway, and one pair of genes in the caffeine biosynthesis pathway, which were expanded and retained following this WGD. Nearly all these gene pairs were expressed in tea plants, implying the contribution of the WGD. This study shows that in addition to the role of the recent tandem gene duplication in the accumulation of tea flavor-related genes, the WGD may have been another main factor driving the evolution of tea flavor.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
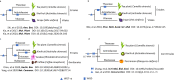
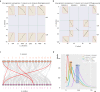
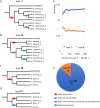
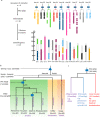

References
-
- Mondal TK, Bhattacharya A, Laxmikumaran M, Ahuja PS. Recent advances of tea (Camellia sinensis) biotechnology. Plant Cell Tissue Organ Cult. 2004;76:195–254. doi: 10.1023/B:TICU.0000009254.87882.71. - DOI

