Molecular characteristics of S-RNase alleles as the determinant of self-incompatibility in the style of Fragaria viridis
- PMID: 34333550
- PMCID: PMC8325692
- DOI: 10.1038/s41438-021-00623-x
Molecular characteristics of S-RNase alleles as the determinant of self-incompatibility in the style of Fragaria viridis
Abstract
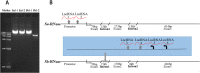
Strawberry (Fragaria spp.) is a member of the Rosoideae subfamily in the family Rosaceae. The self-incompatibility (SI) of some diploid species is a key agronomic trait that acts as a basic pollination barrier; however, the genetic mechanism underlying SI control in strawberry remains unclear. Two candidate S-RNases (Sa- and Sb-RNase) identified in the transcriptome of the styles of the self-incompatible Fragaria viridis 42 were confirmed to be SI determinants at the S locus following genotype identification and intraspecific hybridization using selfing progenies. Whole-genome collinearity and RNase T2 family analysis revealed that only an S locus exists in Fragaria; however, none of the compatible species contained S-RNase. Although the results of interspecific hybridization experiments showed that F. viridis (SI) styles could accept pollen from F. mandshurica (self-compatible), the reciprocal cross was incompatible. Sa and Sb-RNase contain large introns, and their noncoding sequences (promotors and introns) can be transcribed into long noncoding RNAs (lncRNAs). Overall, the genus Fragaria exhibits S-RNase-based gametophytic SI, and S-RNase loss occurs at the S locus of compatible germplasms. In addition, a type of SI-independent unilateral incompatibility exists between compatible and incompatible Fragaria species. Furthermore, the large introns and neighboring lncRNAs in S-RNase in Fragaria could offer clues about S-RNase expression strategies.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







References
-
- Evans W, Jones J. Incompatibility of Fragaria. Can. J. Genet. Cytol. 1967;9:831–836. doi: 10.1139/g67-088. - DOI
-
- Evans W. Evidence of a crossability barrier in diploid x hexaploid and diploid x octoploid crosses in the genus Fragaria. Euphytica. 1974;23:95–100. doi: 10.1007/BF00032746. - DOI
-
- Ahmadi H, Bringhurst R. Breeding strawberries at the decaploid level. J, Amer. Soc. Horti. Sci. 1992;117:856–862. doi: 10.21273/JASHS.117.5.856. - DOI
-
- Marta A, Camadro E, Díaz-Ricci J, Castagnaro A. Breeding barriers between the cultivated strawberry, Fragaria ×ananassa, and related wild germplasm. Euphytica. 2004;136:139. doi: 10.1023/B:EUPH.0000030665.95757.76. - DOI
-
- Bors R, Sullivan J. Interspecific hybridization of F. vesca subspecies with F. nilgerrensis, F. nubicola, F. pentaphylla, and F. viridis. J. Am. Soc. Horticultural Sci. 2005;130:418–423. doi: 10.21273/JASHS.130.3.418. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

