A Metagenome-Wide Association Study of the Gut Microbiome and Metabolic Syndrome
- PMID: 34335505
- PMCID: PMC8322780
- DOI: 10.3389/fmicb.2021.682721
A Metagenome-Wide Association Study of the Gut Microbiome and Metabolic Syndrome
Abstract
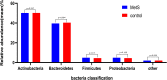
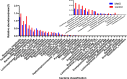
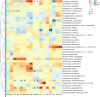
Metabolic syndrome (MetS) is a wide-ranging disorder, which includes insulin resistance, altered glucose and lipid metabolism, and increased blood pressure and visceral obesity. MetS symptoms combine to result in a significant increase in cardiovascular risk. It is therefore critical to treat MetS in the early stages of the disorder. In this study, 123 MetS patients and 304 controls were recruited to determine whether the gut microbiome plays a role in MetS development and progression. By using whole-genome shotgun sequencing, we found that the gut microbiomes of MetS patients were different from those of controls, with MetS patients possessing significantly lower gut microbiome diversity. In addition, 28 bacterial species were negatively correlated with waist circumstance, with Alistipes onderdonkii showing the strongest correlation, followed by Bacteroides thetaiotaomicron, Clostridium asparagiforme, Clostridium citroniae, Clostridium scindens, and Roseburia intestinalis. These species were also enriched in controls relative to MetS patients. In addition, pathways involved in the biosynthesis of carbohydrates, fatty acids, and lipids were enriched in the MetS group, indicating that microbial functions related to fermentation may play a role in MetS. We also found that microbiome changes in MetS patients may aggravate inflammation and contribute to MetS diseases by inhibiting the production of short-chain fatty acids (SCFAs). Taken together, these results indicate the potential utility of beneficial gut microbiota as a potential therapeutic to alleviate MetS.
Keywords: comparative genomics; gut microbiota; metabolic pathway; metabolic syndrome; metagenomics.
Copyright © 2021 Qin, Yan, Yang, Chen, Li, Gao, Yan, Wang, Wang, Wang and Ding.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Bozkurt B., Aguilar D., Deswal A., Dunbar S. B., Francis G. S., Horwich T., et al. (2016). Contributory risk and management of comorbidities of hypertension, obesity, diabetes mellitus, hyperlipidemia, and metabolic syndrome in chronic heart failure: a scientific statement from the American heart association. Circulation 134 e535–e578. 10.1161/CIR.0000000000000450 - DOI - PubMed
LinkOut - more resources
Full Text Sources

